Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
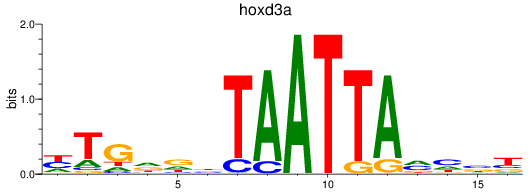
Results for hoxd3a
Z-value: 0.22
Transcription factors associated with hoxd3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd3a
|
ENSDARG00000059280 | homeobox D3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd3a | dr11_v1_chr9_-_1959917_1959917 | -0.48 | 3.6e-02 | Click! |
Activity profile of hoxd3a motif
Sorted Z-values of hoxd3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_35484070 | 0.45 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr19_-_6988837 | 0.38 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr22_+_28320792 | 0.33 |
ENSDART00000160562
ENSDART00000164138 |
impg2b
impg2b
|
interphotoreceptor matrix proteoglycan 2b interphotoreceptor matrix proteoglycan 2b |
| chr23_+_579893 | 0.30 |
ENSDART00000189098
|
BX323461.1
|
|
| chr9_-_21918963 | 0.27 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr18_+_9362455 | 0.27 |
ENSDART00000187025
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr16_+_14707960 | 0.26 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr21_+_8427059 | 0.23 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr22_-_21897203 | 0.22 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr2_-_21819421 | 0.22 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr6_+_3280939 | 0.21 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr8_-_53166975 | 0.20 |
ENSDART00000114683
|
rbsn
|
rabenosyn, RAB effector |
| chr13_-_7233811 | 0.19 |
ENSDART00000162026
|
ninl
|
ninein-like |
| chr14_-_33481428 | 0.19 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr1_+_52392511 | 0.19 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr13_-_44630111 | 0.18 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr14_-_2933185 | 0.17 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr3_-_34084387 | 0.17 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr11_+_13058613 | 0.16 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr20_-_7072487 | 0.15 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr8_-_50888806 | 0.14 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr6_-_57539141 | 0.13 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr1_-_58036509 | 0.13 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr17_-_40956035 | 0.12 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr4_+_47636303 | 0.12 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr4_+_73085993 | 0.11 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr4_+_58576146 | 0.11 |
ENSDART00000164911
|
si:ch211-212k5.4
|
si:ch211-212k5.4 |
| chr5_-_3684402 | 0.11 |
ENSDART00000184354
|
FO704648.1
|
|
| chr8_-_30979494 | 0.11 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr10_+_43039947 | 0.11 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr16_+_45424907 | 0.11 |
ENSDART00000134471
|
phf1
|
PHD finger protein 1 |
| chr11_+_2391469 | 0.10 |
ENSDART00000182121
|
igfbp6a
|
insulin-like growth factor binding protein 6a |
| chr19_-_10971230 | 0.10 |
ENSDART00000166196
|
LO018584.1
|
|
| chr2_-_38284648 | 0.10 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr7_-_12464412 | 0.09 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr19_-_14155781 | 0.09 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr8_+_22405477 | 0.09 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr2_-_16380283 | 0.09 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr16_+_5251768 | 0.08 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr13_-_35808904 | 0.08 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr22_+_29113796 | 0.08 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr17_-_37054959 | 0.07 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr2_+_54327160 | 0.07 |
ENSDART00000182320
|
LO018508.1
|
|
| chr23_-_1348933 | 0.07 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr15_+_5360407 | 0.07 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr7_-_46777876 | 0.07 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr15_+_41027466 | 0.07 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr20_-_9095105 | 0.06 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr8_-_25034411 | 0.06 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr14_-_48961056 | 0.06 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr21_-_35419486 | 0.06 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr16_+_45425181 | 0.05 |
ENSDART00000168591
|
phf1
|
PHD finger protein 1 |
| chr12_+_13405445 | 0.05 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr1_+_56755536 | 0.05 |
ENSDART00000157727
|
CABZ01049361.1
|
|
| chr19_+_2631565 | 0.05 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr3_+_30500968 | 0.05 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr2_-_39558643 | 0.04 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr2_-_28671139 | 0.04 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr19_+_11217279 | 0.04 |
ENSDART00000181859
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr5_+_28160503 | 0.04 |
ENSDART00000051516
|
tacr1a
|
tachykinin receptor 1a |
| chr5_+_66433287 | 0.04 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr5_-_38094130 | 0.04 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr24_+_36204028 | 0.03 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr17_+_23298928 | 0.02 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr10_+_39084354 | 0.02 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr8_+_25034544 | 0.02 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr11_-_25777318 | 0.02 |
ENSDART00000079328
|
plch2b
|
phospholipase C, eta 2b |
| chr20_-_48898560 | 0.02 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr4_-_71210030 | 0.02 |
ENSDART00000186473
|
si:ch211-205a14.7
|
si:ch211-205a14.7 |
| chr18_+_17827149 | 0.02 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr20_-_46362606 | 0.02 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr15_+_31899312 | 0.02 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr14_+_30762131 | 0.02 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr3_+_27798094 | 0.01 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr5_-_28109416 | 0.01 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr6_+_41191482 | 0.01 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr8_-_22274222 | 0.00 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr12_+_30265802 | 0.00 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr10_+_6884627 | 0.00 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.0 | GO:0071314 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.0 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |