Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
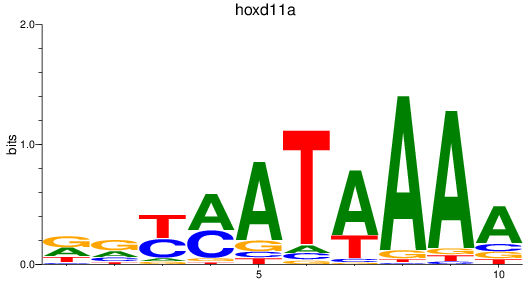
Results for hoxd11a
Z-value: 1.05
Transcription factors associated with hoxd11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd11a
|
ENSDARG00000059267 | homeobox D11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd11a | dr11_v1_chr9_-_1978090_1978090 | -0.83 | 1.2e-05 | Click! |
Activity profile of hoxd11a motif
Sorted Z-values of hoxd11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_40009446 | 6.55 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr23_+_27068225 | 4.99 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr11_+_30244356 | 4.53 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr14_-_33613794 | 3.90 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr24_+_402493 | 3.63 |
ENSDART00000036472
|
VSTM2A
|
zgc:110852 |
| chr7_-_26457208 | 3.44 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr16_-_28658341 | 3.40 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr24_+_40529346 | 3.38 |
ENSDART00000168548
|
CU929259.1
|
|
| chr24_-_6158933 | 3.13 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr2_+_21128391 | 2.94 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr17_-_15498275 | 2.83 |
ENSDART00000156905
ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr4_-_8043839 | 2.74 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr22_+_31821815 | 2.65 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr7_+_2467057 | 2.65 |
ENSDART00000154517
|
si:dkey-125e8.3
|
si:dkey-125e8.3 |
| chr15_+_20403903 | 2.63 |
ENSDART00000134182
|
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr25_+_29161609 | 2.60 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr1_+_17900306 | 2.53 |
ENSDART00000089480
|
cyp4v8
|
cytochrome P450, family 4, subfamily V, polypeptide 8 |
| chr1_+_52481332 | 2.48 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr9_+_33154841 | 2.46 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr6_-_40697585 | 2.38 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr1_-_22512063 | 2.32 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr3_+_49074008 | 2.32 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr24_+_5840258 | 2.31 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr8_+_52637507 | 2.26 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr21_+_11969603 | 2.10 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr2_+_2223837 | 2.02 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr18_+_15876385 | 1.99 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr10_+_2587234 | 1.99 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr18_+_2189211 | 1.96 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr23_-_30431333 | 1.92 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr20_+_25879826 | 1.89 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr7_-_45076131 | 1.87 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr19_+_1370504 | 1.84 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr24_-_30091937 | 1.81 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr16_-_49505275 | 1.79 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr3_+_19299309 | 1.79 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr1_-_14332283 | 1.70 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr25_-_20258508 | 1.70 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr1_+_44173245 | 1.67 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr23_+_11669337 | 1.66 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr10_-_24371312 | 1.66 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr19_+_42469058 | 1.61 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr2_+_5927255 | 1.60 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr25_-_31739309 | 1.60 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr6_-_60147517 | 1.52 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr13_-_36034582 | 1.51 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr2_+_36862473 | 1.50 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr19_+_7810028 | 1.48 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr18_+_38908903 | 1.45 |
ENSDART00000159834
|
myo5aa
|
myosin VAa |
| chr22_+_18786797 | 1.42 |
ENSDART00000141864
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr23_-_12014931 | 1.40 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr25_+_336503 | 1.40 |
ENSDART00000160395
|
CU929262.1
|
|
| chr5_-_42272517 | 1.39 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr1_+_44826593 | 1.37 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr24_+_30392834 | 1.37 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr25_-_13614702 | 1.37 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr24_+_7495945 | 1.37 |
ENSDART00000133525
ENSDART00000182460 ENSDART00000162954 |
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr8_+_40477264 | 1.36 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr10_-_44788554 | 1.36 |
ENSDART00000166965
|
tacr1b
|
tachykinin receptor 1b |
| chr23_+_11669109 | 1.36 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr24_+_7782313 | 1.35 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr18_-_15559817 | 1.35 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr3_+_15271943 | 1.32 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr24_-_11908115 | 1.31 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_-_3366590 | 1.31 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr9_-_23990416 | 1.30 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr3_+_49097775 | 1.27 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr22_+_2207502 | 1.26 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr22_+_8979955 | 1.26 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr25_-_19433244 | 1.25 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr21_-_3770636 | 1.25 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr9_+_54290896 | 1.25 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr1_-_9249943 | 1.24 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr13_-_22843562 | 1.24 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr3_+_29714775 | 1.22 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr21_-_14966718 | 1.19 |
ENSDART00000151200
|
mmp17a
|
matrix metallopeptidase 17a |
| chr3_+_19609638 | 1.17 |
ENSDART00000079319
|
rprml
|
reprimo-like |
| chr22_-_34551568 | 1.17 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr15_+_43166511 | 1.12 |
ENSDART00000011737
|
flj13639
|
flj13639 |
| chr5_+_45140914 | 1.11 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_43585527 | 1.11 |
ENSDART00000161590
|
clrn3
|
clarin 3 |
| chr1_-_9228007 | 1.09 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr9_+_51265283 | 1.09 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr25_-_34973211 | 1.08 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr16_-_52821023 | 1.07 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr18_+_14342326 | 1.02 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr4_+_7391110 | 1.01 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr2_-_43196595 | 1.00 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr2_+_16597011 | 0.99 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr3_-_19495814 | 0.99 |
ENSDART00000162248
|
CU571315.3
|
|
| chr13_-_25719628 | 0.99 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr21_+_27278120 | 0.97 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr21_-_30648106 | 0.97 |
ENSDART00000160800
ENSDART00000177022 |
phka1b
|
phosphorylase kinase, alpha 1b (muscle) |
| chr14_+_45028062 | 0.96 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr11_+_36231248 | 0.95 |
ENSDART00000131104
|
GPR62 (1 of many)
|
si:ch211-213o11.11 |
| chr14_+_36497250 | 0.95 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr20_-_7128612 | 0.93 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr18_-_39188664 | 0.92 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr14_+_15597049 | 0.92 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr24_+_25259154 | 0.91 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr9_-_34986827 | 0.90 |
ENSDART00000137862
|
si:ch211-160b11.4
|
si:ch211-160b11.4 |
| chr20_-_25709247 | 0.90 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr17_+_13031497 | 0.90 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr19_-_617246 | 0.88 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr11_-_36009924 | 0.86 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr3_+_24458899 | 0.85 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr24_+_25258904 | 0.84 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr6_-_33685325 | 0.84 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_-_30734098 | 0.82 |
ENSDART00000133769
|
rp1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr19_-_43750108 | 0.81 |
ENSDART00000171806
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr4_+_19534833 | 0.81 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr13_-_18174864 | 0.79 |
ENSDART00000079884
|
alox5a
|
arachidonate 5-lipoxygenase a |
| chr19_-_32600823 | 0.79 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr6_-_31348999 | 0.79 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr17_+_23554932 | 0.78 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr8_+_7740004 | 0.78 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr25_-_13614863 | 0.78 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr2_-_32738535 | 0.76 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr5_-_62940851 | 0.76 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_+_44711446 | 0.75 |
ENSDART00000193481
ENSDART00000003895 |
ssrp1b
|
structure specific recognition protein 1b |
| chr5_+_25317061 | 0.74 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr10_-_10969596 | 0.74 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr15_+_21276735 | 0.73 |
ENSDART00000111213
|
ubash3bb
|
ubiquitin associated and SH3 domain containing Bb |
| chr23_+_22785375 | 0.71 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr14_-_8271686 | 0.70 |
ENSDART00000165120
|
purab
|
purine-rich element binding protein Ab |
| chr4_+_20954929 | 0.70 |
ENSDART00000143674
|
nav3
|
neuron navigator 3 |
| chr16_+_11188810 | 0.70 |
ENSDART00000186011
|
cicb
|
capicua transcriptional repressor b |
| chr17_-_50355512 | 0.69 |
ENSDART00000041685
|
otofb
|
otoferlin b |
| chr11_-_15090564 | 0.65 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr11_-_40257225 | 0.64 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr4_+_14899720 | 0.64 |
ENSDART00000187456
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr14_-_4120636 | 0.64 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr5_-_57289872 | 0.63 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr16_+_20934353 | 0.62 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr4_+_10888762 | 0.61 |
ENSDART00000136049
|
syt10
|
synaptotagmin X |
| chr15_+_29024895 | 0.61 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr22_+_1421212 | 0.60 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr9_+_30275139 | 0.60 |
ENSDART00000131781
|
rpgra
|
retinitis pigmentosa GTPase regulator a |
| chr20_-_49889111 | 0.60 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr17_-_29311835 | 0.59 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr25_+_33063762 | 0.59 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr14_+_3071110 | 0.59 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr2_+_52847049 | 0.58 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr22_-_9736050 | 0.57 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr22_-_13165186 | 0.57 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr15_-_563877 | 0.55 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr5_+_11290851 | 0.54 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr3_-_19200571 | 0.54 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr11_+_37049347 | 0.53 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr9_-_41784799 | 0.52 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr13_-_37619159 | 0.52 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr16_+_54829293 | 0.52 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr21_+_2102829 | 0.51 |
ENSDART00000157922
|
AL627305.1
|
|
| chr3_-_24456451 | 0.50 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr1_+_2321384 | 0.50 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr17_-_23727978 | 0.50 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr20_+_41906960 | 0.49 |
ENSDART00000193460
|
cep85l
|
centrosomal protein 85, like |
| chr25_-_32449235 | 0.49 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr13_-_25745089 | 0.47 |
ENSDART00000189333
ENSDART00000147420 |
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr12_-_18872717 | 0.46 |
ENSDART00000126300
|
shisa8b
|
shisa family member 8b |
| chr20_-_34028967 | 0.46 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr7_-_52334840 | 0.46 |
ENSDART00000174173
|
CR938716.1
|
|
| chr4_-_22338906 | 0.46 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr15_+_29276508 | 0.44 |
ENSDART00000170537
ENSDART00000126559 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr22_-_32360464 | 0.44 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr23_+_34047413 | 0.44 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr7_-_7692992 | 0.43 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr10_-_42882215 | 0.43 |
ENSDART00000180580
ENSDART00000187374 |
FP236542.1
|
|
| chr5_+_42400777 | 0.43 |
ENSDART00000183114
|
BX548073.8
|
|
| chr7_+_18075504 | 0.42 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr2_-_44038698 | 0.42 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr7_+_13639473 | 0.38 |
ENSDART00000157870
ENSDART00000032899 |
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr20_+_52546186 | 0.37 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr21_-_22827548 | 0.37 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr7_+_24522308 | 0.36 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr15_+_44093286 | 0.36 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr8_-_38159805 | 0.36 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr3_-_29910547 | 0.36 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr24_+_1042594 | 0.35 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr11_-_6265574 | 0.35 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr7_+_66822229 | 0.34 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr4_-_16334362 | 0.34 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr12_-_18872927 | 0.34 |
ENSDART00000187717
|
shisa8b
|
shisa family member 8b |
| chr10_-_36618674 | 0.33 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr10_+_20628698 | 0.33 |
ENSDART00000064666
|
prnpb
|
prion protein b |
| chr7_-_8022741 | 0.33 |
ENSDART00000172841
|
si:ch211-163c2.1
|
si:ch211-163c2.1 |
| chr22_+_2632961 | 0.33 |
ENSDART00000106278
|
znf1173
|
zinc finger protein 1173 |
| chr9_+_13229585 | 0.31 |
ENSDART00000154879
ENSDART00000141705 |
carf
|
calcium responsive transcription factor |
| chr8_+_30709685 | 0.30 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr20_+_43379029 | 0.29 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr11_-_15090118 | 0.28 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr8_+_23521974 | 0.28 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr8_-_43834442 | 0.27 |
ENSDART00000191927
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr8_-_38317914 | 0.26 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr10_-_10969444 | 0.25 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr17_-_2690083 | 0.25 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr3_+_13482542 | 0.24 |
ENSDART00000161287
|
CABZ01079251.1
|
|
| chr8_+_28467893 | 0.24 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr16_+_21242491 | 0.23 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr17_+_52823015 | 0.23 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 1.0 | 3.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.8 | 2.5 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.7 | 6.5 | GO:0006833 | water transport(GO:0006833) |
| 0.5 | 1.4 | GO:0071312 | response to cocaine(GO:0042220) cellular response to alkaloid(GO:0071312) cellular response to cocaine(GO:0071314) |
| 0.4 | 2.6 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.3 | 1.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 1.8 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 2.0 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 1.4 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.2 | 1.4 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.2 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 1.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 1.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.2 | 0.8 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 0.6 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 0.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.1 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.3 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 4.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.1 | 1.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.1 | 0.4 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.4 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 2.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.3 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 2.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.7 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.4 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 2.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 1.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 1.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.6 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 2.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 3.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.7 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 1.7 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 1.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 1.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 2.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 1.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.9 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.4 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 2.1 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 1.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 2.1 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 2.3 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 1.2 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 2.9 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 1.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 1.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.6 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 2.3 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.2 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 1.0 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 1.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 2.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.1 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.3 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 1.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.2 | GO:0030424 | axon(GO:0030424) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.1 | 3.4 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 1.0 | 3.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 2.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.6 | 5.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 1.4 | GO:0016496 | substance P receptor activity(GO:0016496) |
| 0.4 | 1.8 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 2.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.7 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.3 | 1.4 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.2 | 1.0 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.2 | 0.8 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 1.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 3.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 1.5 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 2.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 2.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 2.9 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 2.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.8 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 2.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 1.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.1 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 3.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 2.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 1.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.0 | REACTOME TOLL RECEPTOR CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |