Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
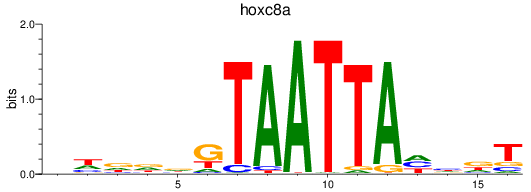
Results for hoxc8a
Z-value: 0.45
Transcription factors associated with hoxc8a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc8a
|
ENSDARG00000070346 | homeobox C8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc8a | dr11_v1_chr23_+_36101185_36101185 | 0.77 | 1.2e-04 | Click! |
Activity profile of hoxc8a motif
Sorted Z-values of hoxc8a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_35808904 | 1.29 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr4_+_5333988 | 1.02 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr16_+_51180938 | 1.00 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr5_+_28497956 | 0.98 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr7_-_48667056 | 0.97 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr11_+_33818179 | 0.75 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr24_+_22485710 | 0.75 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr21_-_26490186 | 0.73 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr14_-_16082806 | 0.70 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr20_-_27225876 | 0.68 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr1_+_19303241 | 0.67 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr8_-_39822917 | 0.64 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr17_+_30369396 | 0.62 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr8_-_31107537 | 0.60 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr1_+_513986 | 0.59 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr4_-_5333971 | 0.57 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr11_-_6877973 | 0.57 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr22_-_16154771 | 0.55 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr2_+_15048410 | 0.53 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr20_+_11731039 | 0.52 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr2_+_16482338 | 0.50 |
ENSDART00000143912
|
fbxo36b
|
F-box protein 36b |
| chr5_+_66433287 | 0.49 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr3_+_22442445 | 0.48 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr2_+_4207209 | 0.47 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr2_+_20793982 | 0.46 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr23_-_16485190 | 0.46 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr1_-_40911332 | 0.46 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr7_+_69019851 | 0.46 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr3_+_30500968 | 0.45 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr25_-_21031007 | 0.44 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr2_-_31833347 | 0.43 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr15_+_841383 | 0.42 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr24_-_38657683 | 0.42 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr20_+_40457599 | 0.41 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr13_+_31321297 | 0.41 |
ENSDART00000143308
|
antxr1d
|
anthrax toxin receptor 1d |
| chr10_+_22034477 | 0.41 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr11_-_21528056 | 0.41 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr24_+_36204028 | 0.41 |
ENSDART00000063832
ENSDART00000155260 |
rbbp8
|
retinoblastoma binding protein 8 |
| chr6_-_43283122 | 0.41 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr15_+_35253543 | 0.41 |
ENSDART00000171673
|
zgc:158328
|
zgc:158328 |
| chr22_+_29113796 | 0.39 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr20_-_45812144 | 0.39 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr6_-_33924883 | 0.39 |
ENSDART00000132762
ENSDART00000148142 ENSDART00000142213 |
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr7_+_29163762 | 0.37 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr21_-_34032650 | 0.37 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr9_+_41080029 | 0.35 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr9_+_32178374 | 0.35 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr11_-_40728380 | 0.33 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr8_-_7567815 | 0.33 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr7_-_30174882 | 0.32 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr5_+_63857055 | 0.31 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr6_-_40713183 | 0.30 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr2_+_3823813 | 0.30 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr21_+_35328025 | 0.29 |
ENSDART00000136211
|
rars
|
arginyl-tRNA synthetase |
| chr13_-_36621926 | 0.29 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr2_-_30668580 | 0.28 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr15_-_43284021 | 0.28 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr21_+_22738939 | 0.28 |
ENSDART00000151342
ENSDART00000079145 |
arhgap42a
|
Rho GTPase activating protein 42a |
| chr10_+_42358426 | 0.28 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr5_+_26213874 | 0.28 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr11_-_1509773 | 0.28 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr1_-_19764038 | 0.27 |
ENSDART00000005733
|
tma16
|
translation machinery associated 16 homolog |
| chr1_-_58036509 | 0.26 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr19_+_10592778 | 0.26 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr1_+_58628165 | 0.25 |
ENSDART00000158654
|
si:ch73-221f6.4
|
si:ch73-221f6.4 |
| chr6_-_55399214 | 0.25 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr19_+_16064439 | 0.25 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr2_-_33687214 | 0.24 |
ENSDART00000147439
|
atp6v0b
|
ATPase H+ transporting V0 subunit b |
| chr18_+_17827149 | 0.24 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr13_-_39736938 | 0.24 |
ENSDART00000141645
|
zgc:171482
|
zgc:171482 |
| chr2_-_36500347 | 0.23 |
ENSDART00000110528
|
BX901889.1
|
|
| chr16_-_13623659 | 0.23 |
ENSDART00000168978
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr19_+_33850705 | 0.23 |
ENSDART00000160356
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr8_-_30204650 | 0.22 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr20_-_29532939 | 0.22 |
ENSDART00000049224
ENSDART00000062377 |
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr19_-_33850201 | 0.22 |
ENSDART00000168583
|
otud6b
|
OTU domain containing 6B |
| chr1_+_22691256 | 0.21 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr1_+_57040472 | 0.21 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr17_+_23298928 | 0.20 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr11_+_11267493 | 0.20 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr15_+_31344472 | 0.19 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr16_+_13818743 | 0.19 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr16_+_4497302 | 0.18 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
| chr25_+_31868268 | 0.18 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr24_+_26997798 | 0.18 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr21_-_32097908 | 0.17 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr10_+_11355841 | 0.17 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr16_-_28878080 | 0.17 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr7_-_32895668 | 0.17 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr11_+_37250839 | 0.16 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr22_+_1170294 | 0.16 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr15_+_29140126 | 0.16 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr10_+_15224660 | 0.16 |
ENSDART00000137932
|
sh3bp2
|
SH3-domain binding protein 2 |
| chr8_+_16676894 | 0.16 |
ENSDART00000076586
|
si:ch211-198n5.11
|
si:ch211-198n5.11 |
| chr4_+_14727212 | 0.16 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr20_+_13403281 | 0.16 |
ENSDART00000045183
ENSDART00000127350 |
enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr2_-_36490570 | 0.15 |
ENSDART00000109828
|
BX901889.3
|
|
| chr21_+_29077509 | 0.15 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr10_+_40568735 | 0.15 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr13_-_46200240 | 0.15 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr10_+_28160265 | 0.15 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr15_+_15856178 | 0.14 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr1_+_524388 | 0.14 |
ENSDART00000020327
|
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr5_+_29671681 | 0.14 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr12_+_48803098 | 0.14 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr22_-_17474781 | 0.14 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr23_+_36130883 | 0.13 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr8_+_31717175 | 0.13 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr13_+_31070181 | 0.13 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr18_+_3037998 | 0.12 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr5_-_29671586 | 0.12 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr15_+_857148 | 0.12 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr11_+_31864921 | 0.12 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr24_-_2450597 | 0.12 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr20_-_7072487 | 0.12 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr25_-_3470910 | 0.12 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr24_+_7322116 | 0.11 |
ENSDART00000005804
|
XRCC2
|
X-ray repair cross complementing 2 |
| chr4_-_22472653 | 0.11 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr5_-_68093169 | 0.11 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr10_+_6884627 | 0.11 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr6_+_19383267 | 0.11 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr18_+_27738349 | 0.10 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr14_-_48961056 | 0.10 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr10_+_518546 | 0.10 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr22_-_17474583 | 0.09 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr11_-_7380674 | 0.08 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
| chr8_+_22516728 | 0.08 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr20_-_49704915 | 0.07 |
ENSDART00000189232
|
COX7A2 (1 of many)
|
cytochrome c oxidase subunit 7A2 |
| chr15_-_37835638 | 0.07 |
ENSDART00000128922
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr8_-_13064330 | 0.07 |
ENSDART00000129168
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr15_-_35252522 | 0.07 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr7_+_4474880 | 0.07 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr23_+_579893 | 0.06 |
ENSDART00000189098
|
BX323461.1
|
|
| chr2_+_3502430 | 0.06 |
ENSDART00000113395
|
SERP1
|
zgc:92744 |
| chr15_-_37104165 | 0.06 |
ENSDART00000165867
|
zmp:0000001114
|
zmp:0000001114 |
| chr20_-_9095105 | 0.06 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr8_-_11004726 | 0.06 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr21_+_76739 | 0.05 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr24_+_12894282 | 0.05 |
ENSDART00000061301
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr5_+_33498253 | 0.05 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr2_+_31833997 | 0.05 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr24_+_16985181 | 0.05 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr20_+_41021054 | 0.05 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr25_-_7705487 | 0.04 |
ENSDART00000128099
|
prdm11
|
PR domain containing 11 |
| chr13_+_35339182 | 0.04 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_-_13623928 | 0.04 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr4_-_11064073 | 0.04 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr12_+_18744610 | 0.04 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr13_+_6219551 | 0.04 |
ENSDART00000025910
|
apmap
|
adipocyte plasma membrane associated protein |
| chr21_+_35327589 | 0.03 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr24_-_25428176 | 0.03 |
ENSDART00000090010
|
phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr18_+_19972853 | 0.03 |
ENSDART00000180071
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr4_+_49506359 | 0.02 |
ENSDART00000154478
|
si:dkey-5i16.5
|
si:dkey-5i16.5 |
| chr4_+_14727018 | 0.02 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr17_+_50248152 | 0.02 |
ENSDART00000153998
|
zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr25_-_13490744 | 0.01 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr21_-_35419486 | 0.01 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr7_-_46777876 | 0.01 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr7_-_45999333 | 0.01 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr2_-_17235891 | 0.00 |
ENSDART00000144251
|
artnb
|
artemin b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc8a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.4 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.6 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.6 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.5 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.5 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.4 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 1.0 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.8 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.5 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.2 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.6 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.6 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.2 | 0.6 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.6 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0030273 | melanin-concentrating hormone receptor activity(GO:0030273) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |