Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
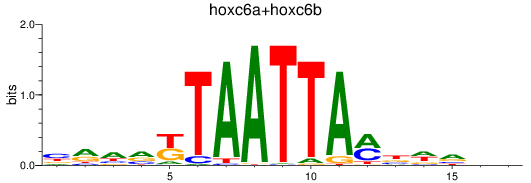
Results for hoxc6a+hoxc6b
Z-value: 0.93
Transcription factors associated with hoxc6a+hoxc6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc6a
|
ENSDARG00000070343 | homeobox C6a |
|
hoxc6b
|
ENSDARG00000101954 | homeobox C6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc6b | dr11_v1_chr11_+_2198831_2198831 | 0.91 | 5.2e-08 | Click! |
| hoxc6a | dr11_v1_chr23_+_36115541_36115541 | 0.06 | 8.2e-01 | Click! |
Activity profile of hoxc6a+hoxc6b motif
Sorted Z-values of hoxc6a+hoxc6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_16422654 | 4.31 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr16_+_54209504 | 3.27 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr3_-_19368435 | 3.03 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr17_+_30369396 | 2.92 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr16_+_29509133 | 2.66 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr25_-_21031007 | 2.56 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr2_-_31833347 | 2.33 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr22_-_16154771 | 2.27 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr2_+_15048410 | 2.20 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr21_-_26490186 | 2.12 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr25_+_22320738 | 1.99 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_+_26534131 | 1.92 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr1_-_22851481 | 1.92 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr23_-_18913032 | 1.89 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr1_+_19303241 | 1.86 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr20_+_54299419 | 1.78 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr24_+_21514283 | 1.74 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr20_+_54309148 | 1.73 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr2_-_10877765 | 1.72 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr1_+_22851261 | 1.70 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr24_-_38657683 | 1.70 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr16_-_21140097 | 1.69 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr7_-_8712148 | 1.68 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr21_+_43172506 | 1.65 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr23_-_16485190 | 1.63 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr17_-_29224908 | 1.61 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr20_+_54304800 | 1.58 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr16_+_28728347 | 1.56 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr20_+_11731039 | 1.56 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr4_+_57881965 | 1.54 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr11_-_10456553 | 1.53 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr6_+_7533601 | 1.51 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr16_+_4654333 | 1.48 |
ENSDART00000167665
|
LIN28A
|
si:ch1073-284b18.2 |
| chr14_-_16082806 | 1.47 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr22_-_36530902 | 1.44 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_-_10456387 | 1.43 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr23_+_1029450 | 1.41 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr1_+_513986 | 1.41 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr7_-_5375214 | 1.39 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr5_+_41477954 | 1.38 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr16_-_7793457 | 1.35 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr22_-_5252005 | 1.34 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr1_-_42289704 | 1.30 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr24_+_34113424 | 1.30 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr5_+_61459422 | 1.25 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr24_+_19415124 | 1.24 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr3_+_40164129 | 1.23 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr11_+_34760628 | 1.20 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr13_+_43247936 | 1.20 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr2_+_10878406 | 1.20 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr5_+_41477526 | 1.18 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr1_-_669717 | 1.18 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr20_+_25586099 | 1.12 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr20_+_40457599 | 1.12 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr13_-_12602920 | 1.08 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr1_+_27977297 | 1.08 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr6_-_43283122 | 1.08 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr5_+_66433287 | 1.05 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr10_+_22527715 | 1.03 |
ENSDART00000134864
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr8_+_26874924 | 1.03 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr20_+_25225112 | 0.97 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr24_+_21540842 | 0.97 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr20_-_45812144 | 0.96 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr10_+_11767791 | 0.95 |
ENSDART00000092047
|
ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr22_-_21676364 | 0.93 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr21_-_26028205 | 0.91 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr14_+_4807207 | 0.91 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr25_-_27722614 | 0.90 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr16_-_27677930 | 0.88 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr23_-_36418708 | 0.88 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
| chr19_+_43359075 | 0.88 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr6_+_35362225 | 0.86 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr8_+_29635968 | 0.85 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr16_-_42965192 | 0.84 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr22_+_508290 | 0.82 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr17_+_41992054 | 0.82 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr25_-_37186894 | 0.81 |
ENSDART00000191647
ENSDART00000182095 |
tdrd12
|
tudor domain containing 12 |
| chr9_-_30576522 | 0.80 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr12_-_28363111 | 0.80 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr6_+_4229360 | 0.79 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr3_+_26342768 | 0.79 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr3_+_38540411 | 0.75 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr21_+_25236297 | 0.75 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr2_+_27010439 | 0.74 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr11_+_17984354 | 0.74 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr19_+_10592778 | 0.73 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr23_+_4709607 | 0.71 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr11_+_11974708 | 0.70 |
ENSDART00000125060
|
zgc:64002
|
zgc:64002 |
| chr5_+_32076109 | 0.70 |
ENSDART00000051357
ENSDART00000144510 |
zmat5
|
zinc finger, matrin-type 5 |
| chr1_-_51038885 | 0.69 |
ENSDART00000035150
|
spast
|
spastin |
| chr15_-_43284021 | 0.69 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr7_+_19495905 | 0.69 |
ENSDART00000125584
ENSDART00000173774 |
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr3_+_28939759 | 0.68 |
ENSDART00000141904
|
lgals1l1
|
lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 |
| chr8_+_50953776 | 0.67 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr7_-_34448076 | 0.67 |
ENSDART00000170935
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr22_-_24791505 | 0.66 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr4_-_43388943 | 0.66 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr21_+_37090585 | 0.65 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr7_+_7696665 | 0.62 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr11_+_37250839 | 0.62 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr7_-_67248829 | 0.61 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr21_-_32097908 | 0.61 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr6_+_25257728 | 0.61 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr2_+_11923615 | 0.60 |
ENSDART00000126118
|
trove2
|
TROVE domain family, member 2 |
| chr8_+_17775247 | 0.59 |
ENSDART00000112356
|
si:ch211-150o23.3
|
si:ch211-150o23.3 |
| chr8_-_30204650 | 0.58 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr20_-_9095105 | 0.58 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr17_+_51682429 | 0.57 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr9_+_41080029 | 0.57 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr11_-_36156935 | 0.57 |
ENSDART00000124935
ENSDART00000138609 |
brk1
gpx1a
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit glutathione peroxidase 1a |
| chr1_+_47499888 | 0.57 |
ENSDART00000027624
|
stn1
|
STN1, CST complex subunit |
| chr9_+_23895711 | 0.56 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr3_+_46635527 | 0.56 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr17_+_20589553 | 0.55 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr2_-_17393216 | 0.54 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr4_-_43640507 | 0.54 |
ENSDART00000150700
|
si:dkey-29p23.2
|
si:dkey-29p23.2 |
| chr15_+_857148 | 0.54 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr24_+_9693951 | 0.52 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr24_+_28953089 | 0.50 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr15_+_29140126 | 0.50 |
ENSDART00000060034
|
zgc:113149
|
zgc:113149 |
| chr9_+_11281969 | 0.50 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr2_+_20406399 | 0.49 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr2_+_2169337 | 0.48 |
ENSDART00000179939
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr13_+_9468535 | 0.47 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr3_-_31158382 | 0.47 |
ENSDART00000076764
ENSDART00000076796 |
smg1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr21_-_1640547 | 0.45 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr22_-_9183944 | 0.45 |
ENSDART00000188599
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr11_+_17984167 | 0.44 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_+_49551614 | 0.43 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr9_-_32177117 | 0.42 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr2_-_17392799 | 0.42 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr6_+_12527725 | 0.40 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr10_-_43771447 | 0.39 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr10_+_29850330 | 0.39 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr7_+_4474880 | 0.39 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr11_+_11267493 | 0.39 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr11_-_40728380 | 0.39 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr7_+_4657987 | 0.39 |
ENSDART00000144321
ENSDART00000142344 |
si:dkey-83f18.2
|
si:dkey-83f18.2 |
| chr10_+_34047352 | 0.39 |
ENSDART00000169333
|
HTRA2 (1 of many)
|
si:dkey-10b15.8 |
| chr8_-_53044300 | 0.38 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_-_16717878 | 0.37 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr22_-_20166660 | 0.37 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr2_-_59327299 | 0.36 |
ENSDART00000133734
|
ftr36
|
finTRIM family, member 36 |
| chr3_+_52545014 | 0.35 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr12_+_23912074 | 0.35 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr4_-_11064073 | 0.35 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr7_+_4694924 | 0.33 |
ENSDART00000144873
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr7_+_66884570 | 0.33 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr4_+_19700308 | 0.33 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr13_+_35528607 | 0.32 |
ENSDART00000075414
ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr4_-_5108844 | 0.32 |
ENSDART00000132666
ENSDART00000136096 |
tmem209
|
transmembrane protein 209 |
| chr17_-_25831569 | 0.31 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr8_+_31717175 | 0.31 |
ENSDART00000013434
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr22_+_2751887 | 0.30 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr14_-_34771371 | 0.30 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr4_+_59845617 | 0.30 |
ENSDART00000167626
ENSDART00000123157 |
si:dkey-196n19.2
|
si:dkey-196n19.2 |
| chr13_+_33268657 | 0.29 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr24_+_16985181 | 0.27 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr18_+_20481982 | 0.26 |
ENSDART00000128139
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr15_-_17619306 | 0.26 |
ENSDART00000184011
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr19_+_46158078 | 0.26 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr16_+_13818743 | 0.25 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr5_+_33498253 | 0.25 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr23_-_19140781 | 0.25 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr9_+_24065855 | 0.24 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr2_+_31833997 | 0.23 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr19_+_1688727 | 0.23 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr7_+_4694762 | 0.23 |
ENSDART00000132862
|
si:ch211-225k7.3
|
si:ch211-225k7.3 |
| chr2_+_19522082 | 0.22 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr2_-_37140423 | 0.22 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr13_-_9467944 | 0.22 |
ENSDART00000136582
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr15_-_2754056 | 0.21 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr22_-_28226948 | 0.21 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr4_-_67980261 | 0.20 |
ENSDART00000182305
|
si:ch211-223k15.1
|
si:ch211-223k15.1 |
| chr7_+_19495379 | 0.20 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr13_+_35339182 | 0.20 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr7_+_66884291 | 0.19 |
ENSDART00000187499
|
sbf2
|
SET binding factor 2 |
| chr4_+_2482046 | 0.18 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr10_-_34046757 | 0.18 |
ENSDART00000099648
|
pimr149
|
Pim proto-oncogene, serine/threonine kinase, related 149 |
| chr23_-_18707418 | 0.17 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr13_+_15656042 | 0.17 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr24_-_40860603 | 0.17 |
ENSDART00000188032
|
CU633479.7
|
|
| chr5_-_51198430 | 0.16 |
ENSDART00000132503
ENSDART00000097473 ENSDART00000165870 |
snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr1_+_58242498 | 0.15 |
ENSDART00000149091
|
ggt1l2.2
|
gamma-glutamyltransferase 1 like 2.2 |
| chr19_+_7847920 | 0.14 |
ENSDART00000132454
|
si:dkeyp-85e10.1
|
si:dkeyp-85e10.1 |
| chr2_+_37140448 | 0.14 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr2_+_19578079 | 0.13 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr7_+_4702269 | 0.12 |
ENSDART00000139880
|
si:ch211-225k7.4
|
si:ch211-225k7.4 |
| chr7_-_46777876 | 0.12 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr20_+_22799641 | 0.12 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr17_+_12285285 | 0.11 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr10_+_31248036 | 0.11 |
ENSDART00000193574
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr20_+_46202188 | 0.11 |
ENSDART00000100523
|
taar13c
|
trace amine associated receptor 13c |
| chr7_+_59020972 | 0.11 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr14_-_7137808 | 0.11 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr5_+_56023186 | 0.11 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr12_-_35944654 | 0.10 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr5_-_66702479 | 0.09 |
ENSDART00000129197
|
mn1b
|
meningioma 1b |
| chr1_+_49668423 | 0.09 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr3_+_23029484 | 0.09 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc6a+hoxc6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 2.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.3 | 1.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 1.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.3 | 1.0 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.3 | 1.4 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.2 | 3.0 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.7 | GO:0051228 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 0.9 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.2 | 0.7 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.2 | 1.3 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.2 | 1.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 1.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.5 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.9 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 2.9 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 1.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.6 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 2.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.9 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.4 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 1.0 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.5 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0045761 | cAMP biosynthetic process(GO:0006171) regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.6 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 1.1 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.2 | GO:0060325 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.3 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 4.0 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 2.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 0.6 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.6 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.8 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.0 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 1.2 | GO:0005795 | Golgi stack(GO:0005795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.7 | 2.0 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 1.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 3.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.3 | 1.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.3 | 1.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.3 | 2.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 2.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 2.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.0 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 1.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 1.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.6 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.2 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 3.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 3.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 2.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 4.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.7 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |