Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
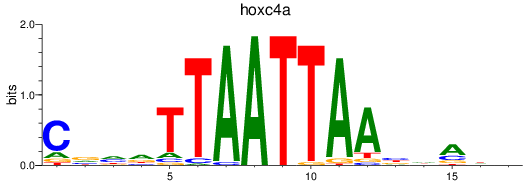
Results for hoxc4a
Z-value: 0.36
Transcription factors associated with hoxc4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc4a
|
ENSDARG00000070338 | homeobox C4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc4a | dr11_v1_chr23_+_36130883_36130883 | 0.45 | 5.2e-02 | Click! |
Activity profile of hoxc4a motif
Sorted Z-values of hoxc4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_21453614 | 0.66 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr17_-_16422654 | 0.66 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr14_+_16287968 | 0.63 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr16_+_54209504 | 0.57 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr24_+_21514283 | 0.53 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr16_-_42894628 | 0.53 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr12_-_35830625 | 0.53 |
ENSDART00000180028
|
CU459056.1
|
|
| chr9_-_1986014 | 0.53 |
ENSDART00000142842
|
hoxd12a
|
homeobox D12a |
| chr1_+_23261857 | 0.52 |
ENSDART00000192774
|
CR759875.5
|
|
| chr1_-_31534089 | 0.51 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr2_+_105748 | 0.49 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr9_-_27398369 | 0.48 |
ENSDART00000186499
|
tex30
|
testis expressed 30 |
| chr5_+_41477954 | 0.47 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr4_+_16715267 | 0.46 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr5_+_41477526 | 0.46 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr25_-_21031007 | 0.41 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr11_+_24815667 | 0.41 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr7_+_38811800 | 0.41 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr4_+_57881965 | 0.41 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr3_+_40164129 | 0.40 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr6_-_33878665 | 0.40 |
ENSDART00000129916
|
tmem69
|
transmembrane protein 69 |
| chr12_-_4243268 | 0.40 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr20_+_40457599 | 0.39 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr1_-_51038885 | 0.38 |
ENSDART00000035150
|
spast
|
spastin |
| chr19_+_10855158 | 0.37 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr20_+_29209615 | 0.36 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_-_26425416 | 0.36 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr19_+_3213914 | 0.36 |
ENSDART00000193144
|
zgc:86598
|
zgc:86598 |
| chr6_+_36821621 | 0.36 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr2_+_38373272 | 0.35 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr22_+_508290 | 0.35 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr2_-_22631498 | 0.34 |
ENSDART00000146425
|
stk25b
|
serine/threonine kinase 25b |
| chr2_+_11685742 | 0.34 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr18_+_41527877 | 0.33 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr14_-_15956921 | 0.33 |
ENSDART00000190500
|
flt4
|
fms-related tyrosine kinase 4 |
| chr23_-_16485190 | 0.33 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr24_+_28953089 | 0.33 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr12_-_31794908 | 0.33 |
ENSDART00000105583
ENSDART00000153449 |
nat9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr23_+_20640484 | 0.32 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr1_+_513986 | 0.32 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr3_+_13848226 | 0.31 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr15_-_34668485 | 0.31 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr8_+_29742237 | 0.31 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr7_-_8712148 | 0.30 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr10_-_31015535 | 0.30 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr24_-_25691020 | 0.30 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr6_-_8392104 | 0.30 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr24_-_38657683 | 0.30 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr22_-_36530902 | 0.29 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr9_+_23895711 | 0.29 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr6_-_40029423 | 0.29 |
ENSDART00000103230
|
pfkfb4b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4b |
| chr3_-_34136778 | 0.28 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr15_-_43284021 | 0.28 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr20_+_48116476 | 0.27 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr1_-_669717 | 0.27 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr20_+_29209767 | 0.27 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr25_+_22320738 | 0.25 |
ENSDART00000073566
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr9_+_37152564 | 0.25 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr3_-_26806032 | 0.25 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr2_+_50608099 | 0.25 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr2_+_20793982 | 0.25 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr13_-_35808904 | 0.24 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr11_+_34760628 | 0.24 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr9_-_14504834 | 0.24 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr20_+_54304800 | 0.24 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr20_+_98179 | 0.23 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr8_+_30664077 | 0.22 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr7_+_21272833 | 0.22 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr20_+_29209926 | 0.22 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr24_+_24461558 | 0.22 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr10_+_16225117 | 0.22 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr6_-_55399214 | 0.22 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr22_-_16042243 | 0.22 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr2_+_36701322 | 0.22 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr17_+_22577472 | 0.21 |
ENSDART00000045099
|
yipf4
|
Yip1 domain family, member 4 |
| chr23_-_17657348 | 0.21 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr12_+_17620067 | 0.21 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr21_+_25236297 | 0.21 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr14_+_22591624 | 0.20 |
ENSDART00000108987
|
gfra4b
|
GDNF family receptor alpha 4b |
| chr20_-_37813863 | 0.20 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr18_-_17485419 | 0.20 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr6_-_35052145 | 0.20 |
ENSDART00000073970
ENSDART00000185790 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr19_-_8877469 | 0.20 |
ENSDART00000193951
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr13_+_48358467 | 0.20 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr23_+_12134839 | 0.20 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr3_+_26288981 | 0.19 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr18_+_14619544 | 0.19 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr3_-_40658820 | 0.19 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr11_+_24800156 | 0.18 |
ENSDART00000131976
|
adipor1a
|
adiponectin receptor 1a |
| chr6_+_25257728 | 0.18 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr24_+_24461341 | 0.18 |
ENSDART00000147658
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_-_45616470 | 0.18 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr24_+_21540842 | 0.18 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr22_-_10156581 | 0.18 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr3_+_36284986 | 0.18 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr10_+_21650828 | 0.17 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr19_+_2631565 | 0.17 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr17_+_41992054 | 0.17 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr4_+_69191065 | 0.17 |
ENSDART00000170595
|
znf1075
|
zinc finger protein 1075 |
| chr11_+_44617021 | 0.17 |
ENSDART00000158887
|
rbm34
|
RNA binding motif protein 34 |
| chr7_+_4474880 | 0.17 |
ENSDART00000143528
|
si:dkey-83f18.14
|
si:dkey-83f18.14 |
| chr19_+_43359075 | 0.17 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr5_-_38451082 | 0.17 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr18_+_7456597 | 0.16 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr23_-_19225709 | 0.16 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr5_+_13394543 | 0.16 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr22_+_737211 | 0.16 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr13_-_12602920 | 0.16 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr25_-_13490744 | 0.16 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr18_+_17827149 | 0.15 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr2_-_7845110 | 0.15 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr23_-_33679579 | 0.15 |
ENSDART00000188674
|
tfcp2
|
transcription factor CP2 |
| chr3_-_29941357 | 0.15 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr10_-_14556978 | 0.15 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr4_+_77943184 | 0.14 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr1_+_6172786 | 0.14 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr2_-_30668580 | 0.14 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr9_+_11281969 | 0.14 |
ENSDART00000110691
|
wnt6b
|
wingless-type MMTV integration site family, member 6b |
| chr22_-_19552796 | 0.14 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr23_+_4709607 | 0.14 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr21_+_27513859 | 0.14 |
ENSDART00000065420
|
pacs1a
|
phosphofurin acidic cluster sorting protein 1a |
| chr3_+_23029484 | 0.14 |
ENSDART00000187900
|
nags
|
N-acetylglutamate synthase |
| chr12_-_19119176 | 0.14 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr4_+_9669717 | 0.13 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr1_+_49668423 | 0.13 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr6_-_11768198 | 0.13 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr6_-_43283122 | 0.13 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr5_+_33498253 | 0.13 |
ENSDART00000140993
|
ms4a17c.2
|
membrane-spanning 4-domains, subfamily A, member 17c.2 |
| chr21_+_25802190 | 0.12 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr16_-_42965192 | 0.12 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr2_+_42318012 | 0.12 |
ENSDART00000138137
|
ftr08
|
finTRIM family, member 8 |
| chr9_-_30576522 | 0.12 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr2_+_10878406 | 0.12 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr20_-_9095105 | 0.12 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr4_+_36150332 | 0.11 |
ENSDART00000161697
|
znf1116
|
zinc finger protein 1116 |
| chr6_-_51771634 | 0.11 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr16_+_23303859 | 0.11 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr6_-_50730749 | 0.11 |
ENSDART00000157153
ENSDART00000110441 |
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr4_-_9891874 | 0.11 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr10_+_26747755 | 0.11 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr5_+_11840905 | 0.10 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr16_+_23796612 | 0.10 |
ENSDART00000131698
|
rab13
|
RAB13, member RAS oncogene family |
| chr8_-_45760087 | 0.10 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr13_-_29420885 | 0.10 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr8_-_53044300 | 0.10 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr2_-_17393216 | 0.10 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr9_-_32177117 | 0.10 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr22_+_5532003 | 0.10 |
ENSDART00000106174
|
si:ch73-256j6.2
|
si:ch73-256j6.2 |
| chr14_+_1170968 | 0.09 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr6_+_15373153 | 0.09 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr17_-_15229787 | 0.09 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr23_+_28809002 | 0.09 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr4_+_33017081 | 0.09 |
ENSDART00000151897
|
si:dkey-26h11.1
|
si:dkey-26h11.1 |
| chr13_-_9450210 | 0.08 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr22_+_26853254 | 0.08 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr6_-_35052388 | 0.08 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr1_+_21731382 | 0.08 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr2_+_37140448 | 0.08 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr18_+_3037998 | 0.08 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr18_+_7456888 | 0.08 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr16_+_24733741 | 0.08 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr9_+_30211038 | 0.07 |
ENSDART00000190847
|
senp7a
|
SUMO1/sentrin specific peptidase 7a |
| chr10_+_29850330 | 0.07 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr2_-_37140423 | 0.07 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr3_+_27606505 | 0.07 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr1_+_51039558 | 0.06 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_+_16625678 | 0.06 |
ENSDART00000164899
|
pcm1
|
pericentriolar material 1 |
| chr24_+_9693951 | 0.06 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr15_+_32798333 | 0.06 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr4_-_13548806 | 0.06 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr17_-_25831569 | 0.06 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr1_+_418869 | 0.06 |
ENSDART00000152173
|
tpp2
|
tripeptidyl peptidase 2 |
| chr19_+_25154066 | 0.06 |
ENSDART00000163220
|
si:ch211-239d6.2
|
si:ch211-239d6.2 |
| chr23_+_20669149 | 0.05 |
ENSDART00000138936
|
arhgef3l
|
Rho guanine nucleotide exchange factor (GEF) 3, like |
| chr9_-_38398789 | 0.05 |
ENSDART00000188384
|
znf142
|
zinc finger protein 142 |
| chr1_+_11882186 | 0.05 |
ENSDART00000193419
|
snx8b
|
sorting nexin 8b |
| chr25_+_13321307 | 0.05 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr19_-_1929310 | 0.05 |
ENSDART00000144113
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr1_+_57787642 | 0.05 |
ENSDART00000127091
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr10_+_44956660 | 0.05 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr2_-_16216568 | 0.05 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr16_+_23431189 | 0.05 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr12_-_47648538 | 0.04 |
ENSDART00000108477
|
fh
|
fumarate hydratase |
| chr5_+_34623107 | 0.04 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr17_-_10122204 | 0.04 |
ENSDART00000160751
|
BX088587.1
|
|
| chr25_+_29474982 | 0.03 |
ENSDART00000130410
|
il17rel
|
interleukin 17 receptor E-like |
| chr10_-_29236860 | 0.03 |
ENSDART00000111620
|
ccdc83
|
coiled-coil domain containing 83 |
| chr7_-_8738827 | 0.03 |
ENSDART00000172807
ENSDART00000173026 |
si:ch211-1o7.3
|
si:ch211-1o7.3 |
| chr25_-_13320986 | 0.03 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr2_+_2223837 | 0.02 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr11_-_2838699 | 0.02 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr22_+_12770877 | 0.02 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr8_-_12867128 | 0.02 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr19_+_11855330 | 0.01 |
ENSDART00000145380
|
adck5
|
aarF domain containing kinase 5 |
| chr1_-_50438247 | 0.01 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr17_+_51682429 | 0.01 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr20_-_31308805 | 0.01 |
ENSDART00000147045
|
hpcal1
|
hippocalcin-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.6 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.4 | GO:0051230 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.3 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 0.4 | GO:0051000 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 0.2 | GO:0090220 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.3 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.3 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.9 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.7 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.0 | 0.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.3 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.9 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.3 | GO:0022829 | wide pore channel activity(GO:0022829) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.7 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |