Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
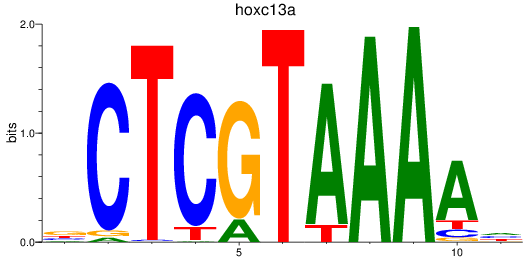
Results for hoxc13a
Z-value: 0.63
Transcription factors associated with hoxc13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc13a
|
ENSDARG00000070353 | homeobox C13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc13a | dr11_v1_chr23_+_36052944_36052944 | -0.27 | 2.7e-01 | Click! |
Activity profile of hoxc13a motif
Sorted Z-values of hoxc13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_22003942 | 3.65 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr25_+_31277415 | 2.03 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr25_+_31276842 | 1.81 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr24_-_26282672 | 1.65 |
ENSDART00000176663
|
and1
|
actinodin1 |
| chr6_+_13779857 | 1.51 |
ENSDART00000154793
|
tmem198b
|
transmembrane protein 198b |
| chr24_-_6158933 | 1.41 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr22_-_16997475 | 1.24 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr5_+_2815021 | 1.21 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr22_-_16997887 | 1.16 |
ENSDART00000138233
|
nfia
|
nuclear factor I/A |
| chr21_+_42226113 | 1.11 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr20_+_23238833 | 1.05 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr12_-_4028079 | 0.86 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr9_-_33749556 | 0.84 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr22_-_24297510 | 0.82 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr4_+_37406676 | 0.78 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr9_-_23990416 | 0.73 |
ENSDART00000113176
|
col6a3
|
collagen, type VI, alpha 3 |
| chr6_+_42338309 | 0.73 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr1_-_40994259 | 0.73 |
ENSDART00000101562
|
adra2c
|
adrenoceptor alpha 2C |
| chr10_-_34867401 | 0.73 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr6_+_49771626 | 0.73 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr3_-_23474416 | 0.72 |
ENSDART00000161672
|
si:dkey-225f5.5
|
si:dkey-225f5.5 |
| chr14_+_24215046 | 0.69 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr11_-_33857911 | 0.68 |
ENSDART00000165370
|
nxph2b
|
neurexophilin 2b |
| chr22_+_18477934 | 0.67 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr1_-_22803147 | 0.66 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr3_+_59851537 | 0.66 |
ENSDART00000180997
|
CU693479.1
|
|
| chr24_+_1294176 | 0.65 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr21_-_37790727 | 0.60 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr14_+_16083818 | 0.60 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr25_+_19947298 | 0.60 |
ENSDART00000067648
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr6_+_49771372 | 0.59 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr25_-_31739309 | 0.58 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr15_-_24883956 | 0.57 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr6_-_54816567 | 0.53 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr15_-_23647078 | 0.52 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr19_-_28367413 | 0.51 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr1_-_22370660 | 0.50 |
ENSDART00000127506
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr15_-_917274 | 0.47 |
ENSDART00000156624
|
si:dkey-77f5.15
|
si:dkey-77f5.15 |
| chr25_-_15214161 | 0.46 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr7_-_31941330 | 0.44 |
ENSDART00000144682
|
bdnf
|
brain-derived neurotrophic factor |
| chr16_-_36099492 | 0.41 |
ENSDART00000180905
|
CU499336.2
|
|
| chr13_-_1349922 | 0.41 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr2_-_20323901 | 0.41 |
ENSDART00000125531
|
plppr5a
|
phospholipid phosphatase related 5a |
| chr23_+_33934228 | 0.40 |
ENSDART00000134237
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr6_-_40098641 | 0.40 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr22_+_24214665 | 0.39 |
ENSDART00000163980
ENSDART00000167996 |
glrx2
|
glutaredoxin 2 |
| chr20_+_51006362 | 0.39 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr9_-_33785093 | 0.38 |
ENSDART00000140779
ENSDART00000059837 |
fundc1
|
FUN14 domain containing 1 |
| chr3_+_35005062 | 0.38 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr22_+_12361317 | 0.38 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr19_+_7506380 | 0.36 |
ENSDART00000182837
|
prcc
|
papillary renal cell carcinoma (translocation-associated) |
| chr16_+_36906693 | 0.35 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr21_-_21594841 | 0.34 |
ENSDART00000132490
|
or133-2
|
odorant receptor, family H, subfamily 133, member 2 |
| chr3_-_7656059 | 0.34 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr9_-_12574473 | 0.33 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr6_+_49412754 | 0.33 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr17_-_3291369 | 0.31 |
ENSDART00000181840
|
CABZ01007222.1
|
|
| chr15_+_16387088 | 0.31 |
ENSDART00000101789
|
flot2b
|
flotillin 2b |
| chr5_-_34993242 | 0.29 |
ENSDART00000134516
ENSDART00000051295 |
btf3
|
basic transcription factor 3 |
| chr7_+_5905091 | 0.28 |
ENSDART00000167099
|
CU459186.3
|
Histone H3.2 |
| chr25_-_3230187 | 0.28 |
ENSDART00000160545
|
cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr23_-_12158685 | 0.28 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr14_+_36497250 | 0.27 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr2_-_56655769 | 0.25 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr5_+_21891305 | 0.24 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr1_+_961607 | 0.24 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr23_+_36115541 | 0.24 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr20_-_2713120 | 0.24 |
ENSDART00000138753
ENSDART00000104606 |
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr9_+_45428041 | 0.24 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr23_+_28582865 | 0.23 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr3_-_36210344 | 0.23 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr19_+_4916233 | 0.19 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr18_+_34225520 | 0.19 |
ENSDART00000126115
|
v2rl1
|
vomeronasal 2 receptor, l1 |
| chr7_-_48263516 | 0.19 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr16_-_26296477 | 0.17 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr16_-_17541890 | 0.17 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr11_-_16093018 | 0.17 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr14_-_7045009 | 0.16 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr12_+_20700961 | 0.16 |
ENSDART00000016099
|
si:ch211-119c20.2
|
si:ch211-119c20.2 |
| chr16_+_22618620 | 0.16 |
ENSDART00000185728
ENSDART00000041625 |
chrnb2b
|
cholinergic receptor, nicotinic, beta 2b |
| chr13_-_10261383 | 0.16 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr2_-_39759059 | 0.15 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr5_-_12031174 | 0.15 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr9_+_12896340 | 0.14 |
ENSDART00000136417
|
si:ch211-167j6.5
|
si:ch211-167j6.5 |
| chr1_+_58282449 | 0.14 |
ENSDART00000131475
|
si:dkey-222h21.7
|
si:dkey-222h21.7 |
| chr25_-_7887274 | 0.13 |
ENSDART00000157276
|
si:ch73-151m17.5
|
si:ch73-151m17.5 |
| chr6_-_19665114 | 0.13 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr19_+_34230108 | 0.12 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr7_+_24114694 | 0.11 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr4_-_2727491 | 0.11 |
ENSDART00000141760
ENSDART00000039083 ENSDART00000134442 |
slco1c1
|
solute carrier organic anion transporter family, member 1C1 |
| chr20_-_27086143 | 0.10 |
ENSDART00000008590
|
itpk1a
|
inositol-tetrakisphosphate 1-kinase a |
| chr1_-_17803614 | 0.10 |
ENSDART00000138475
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr2_+_38881165 | 0.10 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr7_-_23873173 | 0.10 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr4_-_23082985 | 0.09 |
ENSDART00000133644
|
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr18_+_17151916 | 0.09 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr14_-_5546497 | 0.09 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr5_-_31875645 | 0.09 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr5_-_71838520 | 0.08 |
ENSDART00000174396
|
CU927890.1
|
|
| chr4_-_23083187 | 0.08 |
ENSDART00000009768
|
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr23_+_9508538 | 0.08 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr2_+_38147761 | 0.08 |
ENSDART00000135307
|
sall2
|
spalt-like transcription factor 2 |
| chr25_-_13871118 | 0.07 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr25_-_1720736 | 0.07 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr6_+_24661736 | 0.07 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr8_+_18044257 | 0.07 |
ENSDART00000029255
|
glis1b
|
GLIS family zinc finger 1b |
| chr18_+_18405992 | 0.06 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr8_+_14792830 | 0.06 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr6_-_19664848 | 0.06 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr6_+_55285578 | 0.06 |
ENSDART00000180183
|
zgc:109913
|
zgc:109913 |
| chr21_-_25756119 | 0.05 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr9_+_22364997 | 0.05 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr13_-_24447332 | 0.05 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr21_-_21537452 | 0.04 |
ENSDART00000142549
|
or133-7
|
odorant receptor, family H, subfamily 133, member 7 |
| chr4_+_37485430 | 0.04 |
ENSDART00000182667
|
si:dkeyp-86c4.1
|
si:dkeyp-86c4.1 |
| chr13_+_40729618 | 0.04 |
ENSDART00000074829
|
ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr22_-_3299100 | 0.04 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr6_-_8480815 | 0.03 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr21_-_21148623 | 0.03 |
ENSDART00000184364
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr25_+_336503 | 0.03 |
ENSDART00000160395
|
CU929262.1
|
|
| chr20_+_48116476 | 0.02 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr16_+_30933044 | 0.02 |
ENSDART00000184453
|
gstk2
|
glutathione S-transferase kappa 2 |
| chr25_-_3892686 | 0.02 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr21_-_35419486 | 0.01 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr3_+_46764278 | 0.01 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr4_+_42419522 | 0.00 |
ENSDART00000135737
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr17_+_31221761 | 0.00 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 1.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.7 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 0.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.8 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 4.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.0 | 0.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.5 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 3.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 1.7 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.3 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.3 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.3 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 1.2 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 0.5 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 0.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.8 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 3.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.5 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.0 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |