Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
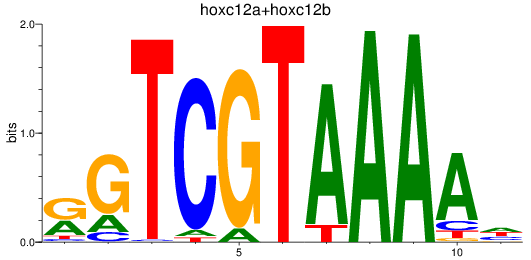
Results for hoxc12a+hoxc12b
Z-value: 1.15
Transcription factors associated with hoxc12a+hoxc12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc12a
|
ENSDARG00000070352 | homeobox C12a |
|
hoxc12b
|
ENSDARG00000103133 | homeobox C12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc12b | dr11_v1_chr11_+_2172335_2172335 | 0.73 | 4.4e-04 | Click! |
| hoxc12a | dr11_v1_chr23_+_36063599_36063599 | 0.36 | 1.3e-01 | Click! |
Activity profile of hoxc12a+hoxc12b motif
Sorted Z-values of hoxc12a+hoxc12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_20715020 | 4.82 |
ENSDART00000015224
|
gadd45gb.1
|
growth arrest and DNA-damage-inducible, gamma b, tandem duplicate 1 |
| chr23_+_36087219 | 3.45 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr12_+_27129659 | 3.22 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr19_+_7424347 | 2.74 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr2_-_17114852 | 2.74 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr11_+_2198831 | 2.62 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr14_+_32022272 | 2.58 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr15_+_29116063 | 2.58 |
ENSDART00000016112
ENSDART00000153609 ENSDART00000155630 |
capns1b
|
calpain, small subunit 1 b |
| chr17_+_21295132 | 2.56 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr3_-_49382896 | 2.50 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr16_+_20910186 | 2.49 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr16_-_6205790 | 2.46 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr19_+_19762183 | 2.39 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr25_+_3294150 | 2.39 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr17_+_17764979 | 2.30 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr21_+_13128180 | 2.22 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr21_-_22357985 | 2.17 |
ENSDART00000101751
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr2_-_6065416 | 2.14 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr5_-_24542726 | 2.02 |
ENSDART00000182975
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr3_+_23710839 | 1.96 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr3_+_23691847 | 1.95 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr23_-_28025943 | 1.86 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr6_+_40775800 | 1.75 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr16_-_12060488 | 1.75 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr2_-_20715094 | 1.73 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr3_+_31058464 | 1.72 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr4_-_13995766 | 1.71 |
ENSDART00000147955
|
prickle1b
|
prickle homolog 1b |
| chr11_-_37717709 | 1.67 |
ENSDART00000177280
|
yod1
|
YOD1 deubiquitinase |
| chr3_-_40254634 | 1.66 |
ENSDART00000154562
|
top3a
|
DNA topoisomerase III alpha |
| chr5_-_31857345 | 1.64 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr7_+_10563017 | 1.63 |
ENSDART00000193520
ENSDART00000173125 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr6_-_9236309 | 1.61 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr9_-_29039506 | 1.61 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr12_-_17199381 | 1.60 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr25_-_7670391 | 1.60 |
ENSDART00000044970
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr22_+_10646928 | 1.58 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr20_+_48803248 | 1.56 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr24_+_21540842 | 1.55 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr2_-_22966076 | 1.55 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr16_+_11242443 | 1.54 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr16_+_5898878 | 1.53 |
ENSDART00000180930
|
ulk4
|
unc-51 like kinase 4 |
| chr1_-_38171648 | 1.51 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr6_-_28943056 | 1.47 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr5_-_31856681 | 1.47 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr5_+_41477954 | 1.46 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr6_+_27992886 | 1.44 |
ENSDART00000160354
|
amotl2a
|
angiomotin like 2a |
| chr16_+_42471455 | 1.42 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr19_+_26640096 | 1.42 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr14_-_26425416 | 1.40 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr19_+_19761966 | 1.39 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr6_-_14038804 | 1.33 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr7_-_30624435 | 1.31 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr13_-_25196758 | 1.30 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr9_+_22634073 | 1.30 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr6_-_53334259 | 1.27 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr16_+_10429770 | 1.25 |
ENSDART00000173132
|
vars
|
valyl-tRNA synthetase |
| chr22_+_10543329 | 1.25 |
ENSDART00000091850
|
atrip
|
ATR interacting protein |
| chr6_-_34838397 | 1.25 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr24_-_24797455 | 1.24 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr12_-_26851726 | 1.24 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr15_-_6863150 | 1.23 |
ENSDART00000153815
|
meis3
|
myeloid ecotropic viral integration site 3 |
| chr7_+_46019780 | 1.23 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr5_-_30715225 | 1.22 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr13_+_29925397 | 1.21 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr23_+_33752275 | 1.21 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr22_+_10440991 | 1.21 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr6_+_27991943 | 1.19 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr19_+_30884960 | 1.18 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr21_-_43131752 | 1.18 |
ENSDART00000024137
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr6_+_3730843 | 1.17 |
ENSDART00000019630
|
FO704755.1
|
|
| chr18_-_31105391 | 1.14 |
ENSDART00000039495
|
pdcd5
|
programmed cell death 5 |
| chr5_-_69499486 | 1.12 |
ENSDART00000023983
ENSDART00000180293 |
psat1
|
phosphoserine aminotransferase 1 |
| chr2_+_31330358 | 1.12 |
ENSDART00000178066
|
clul1
|
clusterin-like 1 (retinal) |
| chr6_-_3992942 | 1.11 |
ENSDART00000182328
|
unc50
|
unc-50 homolog (C. elegans) |
| chr1_-_29061285 | 1.10 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr7_-_18508815 | 1.09 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr19_+_5315987 | 1.09 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr18_+_18000887 | 1.08 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr10_-_31805923 | 1.08 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr11_-_6880725 | 1.08 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr23_+_8797143 | 1.04 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr6_-_53281518 | 1.04 |
ENSDART00000157621
|
rbm5
|
RNA binding motif protein 5 |
| chr12_-_28910419 | 1.03 |
ENSDART00000153278
ENSDART00000152937 |
CCDC189
|
si:ch73-81k8.2 |
| chr9_+_38314466 | 1.00 |
ENSDART00000048753
|
ddx18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr6_+_49551614 | 1.00 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr15_+_5132439 | 1.00 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr3_-_18737126 | 0.99 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr13_-_3936555 | 0.98 |
ENSDART00000017052
|
ncoa4
|
nuclear receptor coactivator 4 |
| chr24_-_31223232 | 0.96 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr3_+_23707691 | 0.96 |
ENSDART00000025449
|
hoxb5a
|
homeobox B5a |
| chr13_-_24447332 | 0.95 |
ENSDART00000043004
|
slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_-_54395488 | 0.94 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr18_-_21748808 | 0.93 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr5_-_26764298 | 0.90 |
ENSDART00000189373
|
rnf181
|
ring finger protein 181 |
| chr18_+_3140682 | 0.90 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr17_+_42274825 | 0.89 |
ENSDART00000020156
|
pax1a
|
paired box 1a |
| chr11_-_36051004 | 0.89 |
ENSDART00000025033
|
gpx1a
|
glutathione peroxidase 1a |
| chr16_-_10316359 | 0.89 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr7_-_25133783 | 0.88 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr18_-_21685055 | 0.88 |
ENSDART00000019861
|
mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr3_-_15734358 | 0.87 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr2_+_23039041 | 0.86 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr22_+_2403068 | 0.86 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr7_+_30240791 | 0.85 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr14_-_32866351 | 0.84 |
ENSDART00000166133
ENSDART00000172545 |
ube2a
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog) |
| chr7_+_29167744 | 0.84 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr2_-_37134169 | 0.83 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr3_+_36284986 | 0.82 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr4_-_28958601 | 0.80 |
ENSDART00000111294
|
zgc:174315
|
zgc:174315 |
| chr14_-_36862745 | 0.77 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr22_-_10440688 | 0.76 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr15_-_28262632 | 0.76 |
ENSDART00000134601
ENSDART00000175022 |
prpf8
|
pre-mRNA processing factor 8 |
| chr3_+_11072958 | 0.75 |
ENSDART00000158899
|
mrps7
|
mitochondrial ribosomal protein S7 |
| chr20_+_34671386 | 0.75 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr8_-_16788626 | 0.72 |
ENSDART00000191652
|
CR759968.2
|
|
| chr10_-_36825984 | 0.72 |
ENSDART00000111104
|
phf12a
|
PHD finger protein 12a |
| chr15_+_27387555 | 0.71 |
ENSDART00000018603
|
tbx4
|
T-box 4 |
| chr4_-_69127091 | 0.71 |
ENSDART00000136092
|
si:ch211-209j12.3
|
si:ch211-209j12.3 |
| chr22_+_17192767 | 0.71 |
ENSDART00000130810
ENSDART00000183006 |
atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr13_+_33268657 | 0.70 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr16_-_47426482 | 0.69 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr5_+_32490238 | 0.68 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr16_-_16761164 | 0.68 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chrM_+_3803 | 0.67 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr12_-_10038870 | 0.67 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr4_-_73488406 | 0.65 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr19_-_33370271 | 0.65 |
ENSDART00000132628
|
nkd3l
|
naked cuticle homolog 3, like |
| chr23_+_23658474 | 0.65 |
ENSDART00000162838
|
agrn
|
agrin |
| chr24_-_24796583 | 0.65 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr20_-_19530751 | 0.65 |
ENSDART00000148574
|
eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr18_+_16943911 | 0.62 |
ENSDART00000157609
|
si:dkey-8l13.5
|
si:dkey-8l13.5 |
| chr22_+_10676981 | 0.61 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr13_+_45431660 | 0.61 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr17_+_23255365 | 0.61 |
ENSDART00000180277
|
AL935174.5
|
|
| chr7_-_23873173 | 0.60 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr12_-_9123352 | 0.59 |
ENSDART00000149655
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr14_-_28052474 | 0.59 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr25_-_7670616 | 0.58 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr13_+_39182099 | 0.58 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr5_-_32489796 | 0.58 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr23_-_39849155 | 0.58 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr20_+_38837238 | 0.58 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr15_-_1590858 | 0.55 |
ENSDART00000081875
|
nnr
|
nanor |
| chr15_+_34675660 | 0.54 |
ENSDART00000009127
|
lta
|
lymphotoxin alpha (TNF superfamily, member 1) |
| chr10_+_26612321 | 0.54 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr21_+_4540127 | 0.53 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr24_+_25913162 | 0.52 |
ENSDART00000143099
ENSDART00000184814 |
map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr8_+_26293673 | 0.51 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr1_-_22834824 | 0.51 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr8_+_23861461 | 0.51 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr2_-_39759059 | 0.50 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr12_+_2677303 | 0.49 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr3_+_24190207 | 0.49 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr3_-_10749691 | 0.47 |
ENSDART00000183088
|
CU210890.1
|
|
| chr11_-_1550709 | 0.46 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr23_+_36115541 | 0.45 |
ENSDART00000130090
|
hoxc6a
|
homeobox C6a |
| chr1_+_31019165 | 0.43 |
ENSDART00000102210
|
itga6b
|
integrin, alpha 6b |
| chr1_+_58424507 | 0.42 |
ENSDART00000114197
|
si:ch73-236c18.3
|
si:ch73-236c18.3 |
| chr20_-_23656516 | 0.42 |
ENSDART00000149735
|
cbr4
|
carbonyl reductase 4 |
| chr16_-_31452416 | 0.41 |
ENSDART00000140880
ENSDART00000008297 ENSDART00000147373 |
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr18_+_33264609 | 0.40 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr16_-_33806390 | 0.37 |
ENSDART00000160671
|
rspo1
|
R-spondin 1 |
| chr18_+_41542542 | 0.37 |
ENSDART00000087445
|
tsen34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr2_+_20406399 | 0.36 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr20_-_23657304 | 0.33 |
ENSDART00000016913
|
cbr4
|
carbonyl reductase 4 |
| chr4_-_30712588 | 0.33 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr2_-_37874647 | 0.32 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr15_-_23942861 | 0.31 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr2_+_20410652 | 0.31 |
ENSDART00000185940
|
palmda
|
palmdelphin a |
| chr11_+_24716837 | 0.31 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr6_-_57476465 | 0.29 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr12_-_30777540 | 0.29 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr1_-_44638058 | 0.29 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr15_-_39820491 | 0.26 |
ENSDART00000097134
|
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_+_13207139 | 0.26 |
ENSDART00000185601
ENSDART00000182182 |
ino80db
|
INO80 complex subunit Db |
| chr7_-_43840418 | 0.25 |
ENSDART00000174592
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr1_+_36612660 | 0.24 |
ENSDART00000190784
|
ednraa
|
endothelin receptor type Aa |
| chr9_-_19161982 | 0.24 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr18_+_7456888 | 0.23 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr11_+_6881001 | 0.22 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr19_+_26718074 | 0.22 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr13_+_8892784 | 0.21 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr16_+_38659475 | 0.20 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr13_-_18119696 | 0.18 |
ENSDART00000148125
|
washc2c
|
WASH complex subunit 2C |
| chr11_-_18557929 | 0.16 |
ENSDART00000110882
ENSDART00000181381 ENSDART00000189312 |
dido1
|
death inducer-obliterator 1 |
| chr5_+_17624463 | 0.16 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr5_-_9216758 | 0.15 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr4_-_38259998 | 0.15 |
ENSDART00000172389
|
znf1093
|
zinc finger protein 1093 |
| chr2_-_55797318 | 0.14 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr24_-_21913426 | 0.13 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr14_-_36863432 | 0.11 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr13_+_21768447 | 0.11 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr15_+_37954666 | 0.10 |
ENSDART00000126534
ENSDART00000155548 |
si:dkey-238d18.7
|
si:dkey-238d18.7 |
| chr24_+_34069675 | 0.10 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr7_+_59739164 | 0.10 |
ENSDART00000110412
|
gpr78b
|
G protein-coupled receptor 78b |
| chr9_-_39005317 | 0.09 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr6_-_28222592 | 0.09 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr3_+_16663373 | 0.07 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr15_-_2754056 | 0.07 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr12_+_46696867 | 0.06 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc12a+hoxc12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.5 | 3.8 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.5 | 4.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.5 | 1.5 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.5 | 2.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.4 | 1.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.4 | 1.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 2.6 | GO:0035912 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.3 | 2.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 0.9 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.3 | 2.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 0.7 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 1.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 1.2 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.2 | 0.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 3.2 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 2.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 3.3 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.2 | 1.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.7 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 1.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 1.2 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.2 | 1.6 | GO:0036372 | opsin transport(GO:0036372) |
| 0.2 | 0.6 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.7 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 0.5 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 2.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.7 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.1 | 1.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.6 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 1.0 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.9 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 1.6 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 2.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.2 | GO:0090220 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.4 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.1 | 1.6 | GO:0072663 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.8 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.9 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 2.2 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 1.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 1.5 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 3.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.7 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.5 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 1.0 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.0 | 2.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 11.4 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 1.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 1.3 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 1.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 1.6 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 1.3 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 1.6 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 2.1 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.3 | 2.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 1.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.3 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 1.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 0.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 0.6 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 2.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.7 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.5 | 2.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 1.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 2.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 2.5 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.3 | 2.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.3 | 1.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 0.8 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 1.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.7 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 0.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.6 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 3.1 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.7 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 1.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 1.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.4 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 1.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 2.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 2.0 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 31.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.2 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.7 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 4.8 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 1.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.7 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 2.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 3.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |