Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
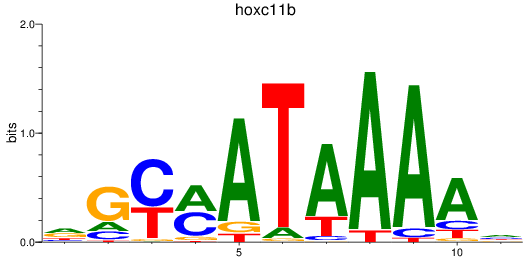
Results for hoxc11b
Z-value: 0.43
Transcription factors associated with hoxc11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11b
|
ENSDARG00000102631 | homeobox C11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc11b | dr11_v1_chr11_+_2180072_2180072 | 0.37 | 1.2e-01 | Click! |
Activity profile of hoxc11b motif
Sorted Z-values of hoxc11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_12588044 | 1.79 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr24_-_40009446 | 1.46 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr7_-_26457208 | 0.95 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr1_+_52481332 | 0.84 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr17_+_27456804 | 0.74 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr24_+_40529346 | 0.68 |
ENSDART00000168548
|
CU929259.1
|
|
| chr1_-_9249943 | 0.68 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr3_+_49074008 | 0.67 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr22_-_31517300 | 0.65 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr21_+_11401247 | 0.64 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr1_+_2321384 | 0.64 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr24_-_29586082 | 0.63 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr2_+_21128391 | 0.63 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr20_-_25533739 | 0.61 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr22_+_31821815 | 0.61 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr24_+_402493 | 0.61 |
ENSDART00000036472
|
VSTM2A
|
zgc:110852 |
| chr22_-_11648322 | 0.61 |
ENSDART00000109596
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr22_-_11648094 | 0.60 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr17_-_2690083 | 0.60 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr14_-_33613794 | 0.60 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr2_-_32738535 | 0.60 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr25_-_31898552 | 0.59 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr25_-_13614702 | 0.56 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr8_+_52637507 | 0.55 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr24_+_5840258 | 0.55 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr24_+_7495945 | 0.54 |
ENSDART00000133525
ENSDART00000182460 ENSDART00000162954 |
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr6_+_30703828 | 0.53 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr2_-_44283554 | 0.52 |
ENSDART00000184684
|
mpz
|
myelin protein zero |
| chr18_+_910992 | 0.52 |
ENSDART00000161206
ENSDART00000167229 |
pkma
|
pyruvate kinase M1/2a |
| chr20_+_25575391 | 0.52 |
ENSDART00000063108
|
cyp2p8
|
cytochrome P450, family 2, subfamily P, polypeptide 8 |
| chr23_+_22785375 | 0.51 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr3_-_3366590 | 0.50 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr15_-_16946124 | 0.48 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr24_+_7782313 | 0.47 |
ENSDART00000111090
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr10_+_2587234 | 0.46 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr12_+_34770531 | 0.45 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr2_+_5927255 | 0.45 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr24_+_30392834 | 0.45 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr9_-_27442339 | 0.45 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr13_+_844150 | 0.44 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr22_-_4644484 | 0.44 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr2_+_52232630 | 0.43 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr16_+_45746549 | 0.43 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr24_-_30091937 | 0.43 |
ENSDART00000148249
|
plppr4b
|
phospholipid phosphatase related 4b |
| chr21_-_39931285 | 0.42 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr2_-_20788698 | 0.42 |
ENSDART00000181823
|
CABZ01020455.1
|
|
| chr2_-_30659222 | 0.42 |
ENSDART00000145405
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr7_-_29534001 | 0.40 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr10_-_36618674 | 0.37 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr14_+_15495088 | 0.37 |
ENSDART00000165765
ENSDART00000188577 |
si:dkey-203a12.6
|
si:dkey-203a12.6 |
| chr12_+_1000323 | 0.37 |
ENSDART00000054363
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr24_-_29822913 | 0.36 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr25_-_13614863 | 0.36 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr25_-_20258508 | 0.36 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr15_+_44093286 | 0.35 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr1_-_22512063 | 0.35 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr7_-_41693004 | 0.35 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr8_-_44299247 | 0.35 |
ENSDART00000144497
|
piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr14_+_49135264 | 0.34 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr2_+_52847049 | 0.34 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr2_+_16597011 | 0.34 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr1_-_23268013 | 0.34 |
ENSDART00000146575
|
rfc1
|
replication factor C (activator 1) 1 |
| chr10_+_44699734 | 0.34 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr5_+_6796291 | 0.34 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr15_-_1843831 | 0.33 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_+_41186320 | 0.33 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr19_+_1370504 | 0.33 |
ENSDART00000158946
|
dgat1a
|
diacylglycerol O-acyltransferase 1a |
| chr18_+_26422124 | 0.32 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr9_+_917060 | 0.32 |
ENSDART00000082390
|
tmem37
|
transmembrane protein 37 |
| chr11_-_40257225 | 0.32 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr15_-_25392589 | 0.31 |
ENSDART00000124205
|
si:dkey-54n8.4
|
si:dkey-54n8.4 |
| chr22_-_34551568 | 0.30 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr10_+_40700311 | 0.30 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr11_+_37049347 | 0.30 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr14_-_1958994 | 0.30 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr1_+_47486104 | 0.30 |
ENSDART00000114746
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr19_-_28367413 | 0.29 |
ENSDART00000079092
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr11_+_37049105 | 0.29 |
ENSDART00000155408
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr25_+_16080181 | 0.28 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
| chr25_-_31739309 | 0.28 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr5_-_28767573 | 0.28 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr12_-_25065916 | 0.28 |
ENSDART00000159167
|
socs5a
|
suppressor of cytokine signaling 5a |
| chr8_-_45867358 | 0.28 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr19_+_7810028 | 0.28 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr12_-_11560794 | 0.28 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr11_-_36009924 | 0.27 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr25_+_336503 | 0.27 |
ENSDART00000160395
|
CU929262.1
|
|
| chr22_+_1911269 | 0.27 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr16_-_49505275 | 0.27 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr8_+_7737062 | 0.27 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr1_+_54683655 | 0.27 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr16_+_4838808 | 0.27 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr19_-_617246 | 0.27 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr23_+_39606108 | 0.27 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr9_-_41784799 | 0.26 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr5_-_57289872 | 0.25 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr9_+_49712868 | 0.25 |
ENSDART00000192969
ENSDART00000183310 |
CSRNP3
|
cysteine and serine rich nuclear protein 3 |
| chr24_-_1356668 | 0.25 |
ENSDART00000188935
|
nrp1a
|
neuropilin 1a |
| chr13_-_25745089 | 0.25 |
ENSDART00000189333
ENSDART00000147420 |
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr24_+_1042594 | 0.24 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr12_-_19091214 | 0.24 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr12_+_27704015 | 0.24 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr18_-_33080454 | 0.23 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr13_-_36034582 | 0.23 |
ENSDART00000133565
|
si:dkey-157l19.2
|
si:dkey-157l19.2 |
| chr10_+_15064433 | 0.23 |
ENSDART00000179978
|
parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr20_+_33532296 | 0.22 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr19_+_42469058 | 0.22 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr2_+_49417900 | 0.22 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr21_-_3700334 | 0.22 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr5_+_45140914 | 0.22 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_7692992 | 0.22 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr2_+_31492662 | 0.22 |
ENSDART00000123495
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr7_+_9981757 | 0.21 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr23_-_26252103 | 0.21 |
ENSDART00000160873
|
magi3a
|
membrane associated guanylate kinase, WW and PDZ domain containing 3a |
| chr16_+_54829293 | 0.21 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr3_-_19200571 | 0.21 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr15_-_16704417 | 0.21 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr10_+_22510280 | 0.21 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr10_+_44700103 | 0.20 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr7_+_38529263 | 0.20 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr6_+_52869892 | 0.20 |
ENSDART00000146143
|
si:dkeyp-3f10.16
|
si:dkeyp-3f10.16 |
| chr16_+_48714048 | 0.20 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr9_-_23156908 | 0.19 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr5_-_67750907 | 0.19 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr2_+_20406399 | 0.19 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr6_-_6448519 | 0.19 |
ENSDART00000180157
ENSDART00000191112 |
si:ch211-194e18.2
|
si:ch211-194e18.2 |
| chr4_+_20954929 | 0.19 |
ENSDART00000143674
|
nav3
|
neuron navigator 3 |
| chr5_-_30074332 | 0.18 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr13_-_15793585 | 0.18 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2 associated athanogene 5 |
| chr4_+_7391400 | 0.18 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr7_+_39634873 | 0.17 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr5_-_42883761 | 0.17 |
ENSDART00000167374
|
BX323596.2
|
|
| chr1_+_8314826 | 0.17 |
ENSDART00000160440
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr24_+_39105051 | 0.17 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr18_-_10713230 | 0.17 |
ENSDART00000183646
|
rbm28
|
RNA binding motif protein 28 |
| chr22_+_2632961 | 0.17 |
ENSDART00000106278
|
znf1173
|
zinc finger protein 1173 |
| chr14_+_3071110 | 0.17 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr25_-_26758253 | 0.17 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr23_-_12014931 | 0.17 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr14_+_15597049 | 0.17 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr25_-_10907786 | 0.17 |
ENSDART00000172490
|
CR339041.1
|
|
| chr10_-_24371312 | 0.16 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr9_+_32859967 | 0.16 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr23_+_2666944 | 0.16 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr18_+_619619 | 0.16 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr6_-_9282080 | 0.16 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr21_+_21906671 | 0.16 |
ENSDART00000016916
|
gria4b
|
glutamate receptor, ionotropic, AMPA 4b |
| chr20_+_52546186 | 0.16 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr24_-_32025637 | 0.16 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr14_-_2221877 | 0.16 |
ENSDART00000106704
|
pcdh2ab1
|
protocadherin 2 alpha b 1 |
| chr4_+_27130412 | 0.16 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr9_+_17306162 | 0.15 |
ENSDART00000075926
|
scel
|
sciellin |
| chr2_-_13254594 | 0.15 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr21_-_17296789 | 0.15 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr15_-_14467394 | 0.15 |
ENSDART00000191944
|
numbl
|
numb homolog (Drosophila)-like |
| chr13_-_319996 | 0.15 |
ENSDART00000148675
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr7_-_7692723 | 0.15 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr24_+_21720304 | 0.15 |
ENSDART00000147250
ENSDART00000048273 |
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr18_-_15373620 | 0.15 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr24_+_26134029 | 0.14 |
ENSDART00000185134
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr3_+_49097775 | 0.14 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr20_-_51831657 | 0.14 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr20_+_40237441 | 0.14 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr2_+_36862473 | 0.13 |
ENSDART00000135624
|
si:dkey-193b15.8
|
si:dkey-193b15.8 |
| chr2_-_43196595 | 0.13 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr14_-_36763302 | 0.13 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr18_+_14342326 | 0.13 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr9_-_12886108 | 0.13 |
ENSDART00000177283
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr15_-_563877 | 0.12 |
ENSDART00000128032
|
cbln18
|
cerebellin 18 |
| chr23_+_18722915 | 0.12 |
ENSDART00000025057
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr5_-_57723929 | 0.12 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr12_-_18872717 | 0.12 |
ENSDART00000126300
|
shisa8b
|
shisa family member 8b |
| chr11_-_42730063 | 0.12 |
ENSDART00000169776
|
si:ch73-106k19.2
|
si:ch73-106k19.2 |
| chr2_+_22409249 | 0.12 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr3_-_58798377 | 0.12 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr10_+_16069987 | 0.11 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr6_+_4528631 | 0.11 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr8_+_30709685 | 0.11 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr8_-_38159805 | 0.11 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr21_+_9689103 | 0.11 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr2_-_44038698 | 0.11 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr7_-_71384391 | 0.11 |
ENSDART00000112841
|
ccdc149a
|
coiled-coil domain containing 149a |
| chr24_-_11908115 | 0.10 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr12_-_44759246 | 0.10 |
ENSDART00000162888
|
dock1
|
dedicator of cytokinesis 1 |
| chr8_-_28655338 | 0.10 |
ENSDART00000017440
|
mc3r
|
melanocortin 3 receptor |
| chr11_+_25112269 | 0.10 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr3_+_24134418 | 0.10 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr24_+_25258904 | 0.10 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr20_-_4883673 | 0.10 |
ENSDART00000145540
ENSDART00000053877 |
zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr14_-_3229384 | 0.10 |
ENSDART00000167247
|
pdgfrb
|
platelet-derived growth factor receptor, beta polypeptide |
| chr8_+_11687254 | 0.09 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr16_+_20934353 | 0.09 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr22_+_19111444 | 0.09 |
ENSDART00000109655
|
fgf22
|
fibroblast growth factor 22 |
| chr2_-_59178742 | 0.09 |
ENSDART00000170594
ENSDART00000109246 |
ftr95
|
finTRIM family, member 95 |
| chr17_-_8592824 | 0.09 |
ENSDART00000127022
|
CU462878.1
|
|
| chr6_+_39114345 | 0.09 |
ENSDART00000136835
|
gpr84
|
G protein-coupled receptor 84 |
| chr12_+_1000526 | 0.09 |
ENSDART00000182556
|
si:ch1073-272o11.3
|
si:ch1073-272o11.3 |
| chr3_+_13482542 | 0.09 |
ENSDART00000161287
|
CABZ01079251.1
|
|
| chr20_+_19212962 | 0.09 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr15_+_32405959 | 0.08 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 1.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.4 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.4 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.4 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.6 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.6 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.1 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 0.3 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.5 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.7 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.2 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.8 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.0 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.4 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.3 | 1.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 1.8 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 0.6 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.4 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.3 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.4 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.4 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.9 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.6 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |