Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
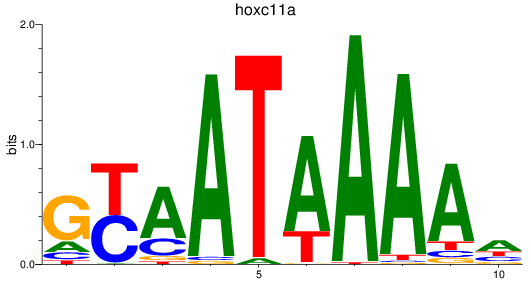
Results for hoxc11a
Z-value: 0.83
Transcription factors associated with hoxc11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11a
|
ENSDARG00000070351 | homeobox C11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc11a | dr11_v1_chr23_+_36074798_36074880 | -0.34 | 1.6e-01 | Click! |
Activity profile of hoxc11a motif
Sorted Z-values of hoxc11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_27068225 | 3.47 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr1_+_44439661 | 3.03 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr2_+_2223837 | 2.80 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr25_+_29161609 | 2.59 |
ENSDART00000180752
|
pkmb
|
pyruvate kinase M1/2b |
| chr23_-_27571667 | 2.29 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr3_+_25154078 | 2.19 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr17_-_12389259 | 2.07 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr10_-_29903165 | 2.05 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr11_+_30244356 | 2.03 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr6_-_60147517 | 1.85 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr25_-_26758253 | 1.75 |
ENSDART00000123004
|
si:dkeyp-73b11.8
|
si:dkeyp-73b11.8 |
| chr7_+_39386982 | 1.74 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr6_-_40697585 | 1.62 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr20_-_27733683 | 1.50 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr12_-_25916530 | 1.46 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr24_-_6158933 | 1.43 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr14_+_49135264 | 1.41 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr3_+_29714775 | 1.34 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr8_-_52413032 | 1.31 |
ENSDART00000183039
|
CABZ01070469.1
|
|
| chr20_-_47731768 | 1.28 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr14_-_33613794 | 1.24 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr8_-_34052019 | 1.17 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr9_+_54290896 | 1.12 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr2_-_27329214 | 1.12 |
ENSDART00000145835
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr20_+_30490682 | 1.10 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr6_-_26559921 | 1.08 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr1_-_22512063 | 1.06 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr23_-_12014931 | 1.04 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr11_+_21053488 | 1.04 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr5_-_42272517 | 1.02 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr2_-_27329667 | 1.02 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr1_+_36437585 | 1.01 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr20_-_25709247 | 0.98 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr18_-_14941840 | 0.97 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr23_-_30431333 | 0.95 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr6_+_12853655 | 0.95 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr9_+_34425736 | 0.92 |
ENSDART00000135147
|
si:ch211-218d20.15
|
si:ch211-218d20.15 |
| chr17_-_30975707 | 0.90 |
ENSDART00000138346
|
evla
|
Enah/Vasp-like a |
| chr19_-_28789404 | 0.90 |
ENSDART00000191453
ENSDART00000026992 |
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr7_+_25059845 | 0.86 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr25_+_20089986 | 0.86 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr22_-_17052381 | 0.82 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr22_-_14115292 | 0.82 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr19_-_47832853 | 0.81 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr14_-_12837432 | 0.79 |
ENSDART00000178444
|
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr22_-_16377960 | 0.78 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr6_-_50203682 | 0.74 |
ENSDART00000083999
ENSDART00000143050 |
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr10_+_36650222 | 0.74 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr17_-_19019635 | 0.73 |
ENSDART00000126666
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr14_-_12837052 | 0.72 |
ENSDART00000165004
ENSDART00000043180 |
gria3b
|
glutamate receptor, ionotropic, AMPA 3b |
| chr11_-_15090564 | 0.71 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr19_-_10243148 | 0.70 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr16_+_10918252 | 0.69 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr11_-_6188413 | 0.68 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr5_+_36611128 | 0.66 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr17_+_52823015 | 0.66 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr11_-_6265574 | 0.66 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr19_+_7810028 | 0.66 |
ENSDART00000081592
ENSDART00000140719 |
aqp10b
|
aquaporin 10b |
| chr5_-_63218919 | 0.64 |
ENSDART00000149979
|
tecta
|
tectorin alpha |
| chr24_+_5840258 | 0.63 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr23_+_11669109 | 0.63 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr18_-_15559817 | 0.63 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr16_-_27749172 | 0.62 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr23_+_28648864 | 0.61 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr23_+_11669337 | 0.60 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr24_+_25259154 | 0.60 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr24_-_29822913 | 0.60 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr11_-_15090118 | 0.60 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr5_+_24245682 | 0.60 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr17_-_47090440 | 0.59 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr17_-_30975978 | 0.59 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr16_+_32995882 | 0.59 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr7_-_29341233 | 0.58 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr11_+_14284866 | 0.57 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr2_-_45663945 | 0.57 |
ENSDART00000075080
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr10_-_34772211 | 0.56 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr20_-_45060241 | 0.54 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr6_-_28222592 | 0.53 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr2_+_21128391 | 0.53 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr23_-_7799184 | 0.52 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr5_+_20147830 | 0.52 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr11_+_24313931 | 0.51 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr25_-_34973211 | 0.51 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr18_-_42333428 | 0.49 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr12_+_20693743 | 0.49 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr24_+_10202718 | 0.48 |
ENSDART00000126668
|
pou6f2
|
POU class 6 homeobox 2 |
| chr3_-_29910547 | 0.47 |
ENSDART00000151501
|
RUNDC1
|
si:dkey-151m15.5 |
| chr24_+_40529346 | 0.47 |
ENSDART00000168548
|
CU929259.1
|
|
| chr1_-_39976492 | 0.46 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr17_+_52822422 | 0.46 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr6_-_58910402 | 0.45 |
ENSDART00000156662
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr1_-_19215336 | 0.43 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr22_-_34551568 | 0.42 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr22_-_13165186 | 0.41 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr16_-_26074529 | 0.41 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr6_+_23931236 | 0.41 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr6_+_52804267 | 0.40 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr19_+_42469058 | 0.40 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr6_-_37468971 | 0.39 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr5_-_30074332 | 0.38 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr21_+_27278120 | 0.38 |
ENSDART00000193882
|
si:dkey-175m17.7
|
si:dkey-175m17.7 |
| chr7_+_30875273 | 0.38 |
ENSDART00000173693
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr11_+_24314148 | 0.38 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_+_2587234 | 0.38 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr19_+_19652439 | 0.37 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr6_-_12588044 | 0.37 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr24_+_26134029 | 0.37 |
ENSDART00000185134
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr1_+_44826593 | 0.36 |
ENSDART00000162200
|
STX3
|
zgc:165520 |
| chr25_+_37126921 | 0.36 |
ENSDART00000124331
|
si:ch1073-174d20.1
|
si:ch1073-174d20.1 |
| chr17_+_52822831 | 0.36 |
ENSDART00000193368
|
meis2a
|
Meis homeobox 2a |
| chr15_-_33896159 | 0.35 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr10_+_35526528 | 0.35 |
ENSDART00000184110
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr14_-_11507211 | 0.35 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr7_+_38529263 | 0.34 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr25_+_336503 | 0.34 |
ENSDART00000160395
|
CU929262.1
|
|
| chr18_+_16330025 | 0.34 |
ENSDART00000142353
|
nts
|
neurotensin |
| chr2_-_43196595 | 0.33 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr24_+_7495945 | 0.32 |
ENSDART00000133525
ENSDART00000182460 ENSDART00000162954 |
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr16_-_35329803 | 0.32 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr24_+_25258904 | 0.32 |
ENSDART00000155714
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr22_+_31821815 | 0.31 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr15_+_14856307 | 0.31 |
ENSDART00000167213
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr2_+_45068366 | 0.30 |
ENSDART00000142175
|
si:dkey-76d14.2
|
si:dkey-76d14.2 |
| chr8_-_18899427 | 0.30 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr13_-_25719628 | 0.30 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr1_-_9228007 | 0.28 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr18_+_48446704 | 0.28 |
ENSDART00000134817
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr13_-_4018888 | 0.28 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr11_+_12052791 | 0.27 |
ENSDART00000158479
|
si:ch211-156l18.8
|
si:ch211-156l18.8 |
| chr23_+_13124085 | 0.27 |
ENSDART00000139475
|
samd10b
|
sterile alpha motif domain containing 10b |
| chr2_-_4925025 | 0.27 |
ENSDART00000160257
|
dlg1l
|
discs, large (Drosophila) homolog 1, like |
| chr8_+_3379815 | 0.26 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr11_-_12998400 | 0.26 |
ENSDART00000018614
|
chrna4b
|
cholinergic receptor, nicotinic, alpha 4b |
| chr11_-_25539323 | 0.26 |
ENSDART00000155785
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr15_-_25209308 | 0.26 |
ENSDART00000157857
|
mnta
|
MAX network transcriptional repressor a |
| chr20_-_33961697 | 0.24 |
ENSDART00000061765
|
selp
|
selectin P |
| chr12_-_46959990 | 0.24 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr2_+_5927255 | 0.24 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr11_-_37589293 | 0.23 |
ENSDART00000172989
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr7_-_26457208 | 0.23 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr5_+_31946973 | 0.23 |
ENSDART00000189876
ENSDART00000163366 |
ungb
|
uracil DNA glycosylase b |
| chr16_+_13883872 | 0.23 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr3_+_52545014 | 0.22 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr21_-_26520629 | 0.22 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr3_-_18675688 | 0.22 |
ENSDART00000048218
|
slc5a11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr2_+_30960351 | 0.22 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr1_-_52447364 | 0.22 |
ENSDART00000140740
|
si:ch211-217k17.10
|
si:ch211-217k17.10 |
| chr9_-_11550711 | 0.22 |
ENSDART00000093343
|
fev
|
FEV (ETS oncogene family) |
| chr16_+_28578648 | 0.21 |
ENSDART00000149566
|
nmt2
|
N-myristoyltransferase 2 |
| chr16_-_21038015 | 0.21 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr16_-_16225260 | 0.20 |
ENSDART00000165790
|
gra
|
granulito |
| chr7_+_18075504 | 0.20 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr14_-_15171435 | 0.20 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr24_+_30392834 | 0.20 |
ENSDART00000162555
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr15_-_13254480 | 0.20 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr17_-_26721007 | 0.19 |
ENSDART00000034580
|
calm1a
|
calmodulin 1a |
| chr5_-_37881345 | 0.19 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr13_+_22675802 | 0.19 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr24_-_8732519 | 0.19 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr8_-_25327809 | 0.18 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr10_+_26612321 | 0.18 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr3_-_3366590 | 0.18 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr16_+_44768361 | 0.17 |
ENSDART00000036302
|
upk1a
|
uroplakin 1a |
| chr15_+_32643873 | 0.17 |
ENSDART00000189433
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr5_+_58687541 | 0.17 |
ENSDART00000083015
ENSDART00000181902 |
ccdc84
|
coiled-coil domain containing 84 |
| chr1_+_45217425 | 0.17 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr17_+_47090497 | 0.17 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr3_-_19200571 | 0.16 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr2_-_17235891 | 0.15 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr21_-_3700334 | 0.15 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr20_+_43925266 | 0.15 |
ENSDART00000037379
|
clic5b
|
chloride intracellular channel 5b |
| chr15_+_31816835 | 0.15 |
ENSDART00000189658
ENSDART00000186634 ENSDART00000193032 ENSDART00000180401 |
frya
|
furry homolog a (Drosophila) |
| chr14_+_15191176 | 0.14 |
ENSDART00000183447
ENSDART00000193093 ENSDART00000169309 |
si:dkey-203a12.2
|
si:dkey-203a12.2 |
| chr17_+_31221761 | 0.14 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr9_+_27354653 | 0.13 |
ENSDART00000134134
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr2_+_14992879 | 0.13 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr7_+_29080684 | 0.13 |
ENSDART00000173709
ENSDART00000173576 |
acd
|
ACD, shelterin complex subunit and telomerase recruitment factor |
| chr19_+_21362553 | 0.13 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr22_-_9736050 | 0.13 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr21_+_20396858 | 0.12 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr14_+_23184517 | 0.12 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr10_+_40700311 | 0.11 |
ENSDART00000157650
ENSDART00000138342 |
taar19n
|
trace amine associated receptor 19n |
| chr21_+_16980141 | 0.11 |
ENSDART00000101241
|
aqp3b
|
aquaporin 3b |
| chr1_+_27153859 | 0.11 |
ENSDART00000180184
|
bnc2
|
basonuclin 2 |
| chr16_+_20934353 | 0.11 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr12_+_19356623 | 0.11 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr7_-_29223614 | 0.10 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr1_+_961607 | 0.10 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr7_-_13750108 | 0.09 |
ENSDART00000173014
|
kbtbd13
|
kelch repeat and BTB (POZ) domain containing 13 |
| chr5_-_57723929 | 0.09 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr11_+_41243325 | 0.09 |
ENSDART00000170657
|
pax7a
|
paired box 7a |
| chr11_-_36009924 | 0.09 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr10_+_40583930 | 0.09 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr20_+_26880668 | 0.09 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr24_+_26134209 | 0.09 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr5_+_33519943 | 0.09 |
ENSDART00000131316
|
ms4a17c.1
|
membrane-spanning 4-domains, subfamily A, member 17C.1 |
| chr3_+_23731109 | 0.08 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr15_-_19772372 | 0.08 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr9_+_917060 | 0.08 |
ENSDART00000082390
|
tmem37
|
transmembrane protein 37 |
| chr19_+_19761966 | 0.08 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.5 | 1.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.5 | 4.1 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.9 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 0.7 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.2 | 2.3 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 0.9 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.6 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 2.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 2.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.8 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.6 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 1.0 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.2 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.0 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 1.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.1 | 2.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 2.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.6 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.2 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.2 | GO:0019860 | thymine catabolic process(GO:0006210) uracil catabolic process(GO:0006212) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.4 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.0 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 2.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.3 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.0 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.5 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 2.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.9 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 3.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 1.5 | GO:0044306 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.6 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.5 | 1.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 2.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 2.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 3.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 2.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.3 | 0.8 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 2.3 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 0.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.6 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.6 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 1.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.9 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.1 | 0.8 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 2.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 5.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 2.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 1.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |