Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
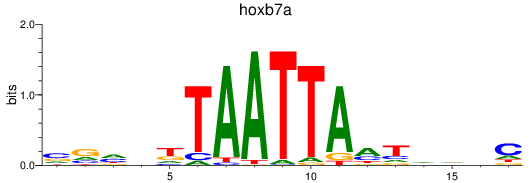
Results for hoxb7a
Z-value: 0.47
Transcription factors associated with hoxb7a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb7a
|
ENSDARG00000056030 | homeobox B7a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb7a | dr11_v1_chr3_+_23691847_23691847 | -0.25 | 3.1e-01 | Click! |
Activity profile of hoxb7a motif
Sorted Z-values of hoxb7a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_1139378 | 1.02 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr3_-_31079186 | 0.73 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr5_-_30620625 | 0.60 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr15_-_40246396 | 0.58 |
ENSDART00000063777
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr25_-_35599887 | 0.52 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr4_-_1801519 | 0.51 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr25_-_19420949 | 0.46 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr12_+_16168342 | 0.45 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr4_-_2545310 | 0.45 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr15_+_28318005 | 0.45 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr16_-_12173399 | 0.44 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr2_-_54039293 | 0.43 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr11_-_45141309 | 0.41 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_+_705727 | 0.38 |
ENSDART00000165366
|
timp4.2
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 2 |
| chr3_-_36602379 | 0.38 |
ENSDART00000161501
ENSDART00000162396 |
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr19_+_5480327 | 0.37 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr6_-_43677125 | 0.37 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr21_-_13123176 | 0.37 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr13_-_5252559 | 0.37 |
ENSDART00000181652
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr22_-_19102256 | 0.34 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr6_+_24398907 | 0.33 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr17_+_30369396 | 0.33 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr12_+_47698356 | 0.33 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr8_+_7801060 | 0.31 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr25_+_14087045 | 0.30 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr18_+_45781516 | 0.30 |
ENSDART00000168840
|
rpl35a
|
ribosomal protein L35a |
| chr14_-_4556896 | 0.29 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr24_+_7800486 | 0.28 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr10_-_13343831 | 0.26 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr18_+_2228737 | 0.25 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr17_+_44697604 | 0.24 |
ENSDART00000156625
|
pgfb
|
placental growth factor b |
| chr5_-_16218777 | 0.24 |
ENSDART00000141698
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr21_+_34088110 | 0.23 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr17_+_31914877 | 0.22 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr15_+_857148 | 0.22 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr16_+_40024883 | 0.22 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr13_-_10945288 | 0.22 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr4_-_63165848 | 0.21 |
ENSDART00000183789
|
CR450780.1
|
|
| chr13_+_10945337 | 0.20 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr15_-_14552101 | 0.20 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr18_+_45781115 | 0.19 |
ENSDART00000151699
ENSDART00000179887 |
rpl35a
|
ribosomal protein L35a |
| chr3_-_50443607 | 0.18 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr12_+_20641102 | 0.17 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr25_+_13620555 | 0.17 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr10_+_11261576 | 0.17 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr21_+_8276989 | 0.17 |
ENSDART00000028265
|
nr5a1b
|
nuclear receptor subfamily 5, group A, member 1b |
| chr12_+_31735159 | 0.17 |
ENSDART00000185442
|
RNF157
|
si:dkey-49c17.3 |
| chr9_+_54039006 | 0.16 |
ENSDART00000112441
|
tlr7
|
toll-like receptor 7 |
| chr23_+_39461989 | 0.16 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr7_+_7696665 | 0.16 |
ENSDART00000091099
|
ino80b
|
INO80 complex subunit B |
| chr5_+_71802014 | 0.15 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr18_-_33080454 | 0.15 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr5_-_67750907 | 0.15 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr23_+_39462208 | 0.15 |
ENSDART00000136707
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_-_1486023 | 0.14 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr18_+_14619544 | 0.14 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr18_+_34362608 | 0.14 |
ENSDART00000131478
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr12_+_48803098 | 0.13 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr22_-_22301672 | 0.12 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr8_-_12867434 | 0.12 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr17_+_51682429 | 0.12 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr7_+_69019851 | 0.11 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr8_-_12867128 | 0.11 |
ENSDART00000142201
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
| chr1_+_19303241 | 0.10 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr6_+_21992820 | 0.10 |
ENSDART00000147507
|
thumpd3
|
THUMP domain containing 3 |
| chr8_+_25034544 | 0.09 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr4_-_73739119 | 0.09 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr23_-_36857964 | 0.09 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr4_+_70151160 | 0.08 |
ENSDART00000111816
|
si:dkey-3h2.4
|
si:dkey-3h2.4 |
| chr5_-_23675222 | 0.08 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr4_+_77971104 | 0.08 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr10_+_43039947 | 0.07 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr13_+_12299997 | 0.07 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr17_+_25481466 | 0.06 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr23_+_28809002 | 0.06 |
ENSDART00000134121
ENSDART00000183661 |
pex14
|
peroxisomal biogenesis factor 14 |
| chr19_+_2631565 | 0.06 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr10_-_11261565 | 0.05 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr24_-_36680261 | 0.05 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr5_+_34623107 | 0.04 |
ENSDART00000184126
|
enc1
|
ectodermal-neural cortex 1 |
| chr3_+_27798094 | 0.03 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr13_-_2440622 | 0.03 |
ENSDART00000102767
ENSDART00000172616 |
fbxo9
|
F-box protein 9 |
| chr17_+_16090436 | 0.03 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr12_+_20641471 | 0.03 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr18_-_15551360 | 0.02 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr22_-_27296889 | 0.01 |
ENSDART00000155724
|
si:dkey-208m12.3
|
si:dkey-208m12.3 |
| chr23_+_4709607 | 0.01 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb7a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 1.0 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.5 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.5 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0048903 | anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.5 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.2 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.7 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.5 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.5 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |