Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for hoxb6a+hoxb6b
Z-value: 3.86
Transcription factors associated with hoxb6a+hoxb6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb6a
|
ENSDARG00000010630 | homeobox B6a |
|
hoxb6b
|
ENSDARG00000026513 | homeobox B6b |
|
hoxb6b
|
ENSDARG00000111786 | homeobox B6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb6a | dr11_v1_chr3_+_23703704_23703704 | -0.50 | 3.1e-02 | Click! |
| hoxb6b | dr11_v1_chr12_+_27127139_27127139 | 0.13 | 5.9e-01 | Click! |
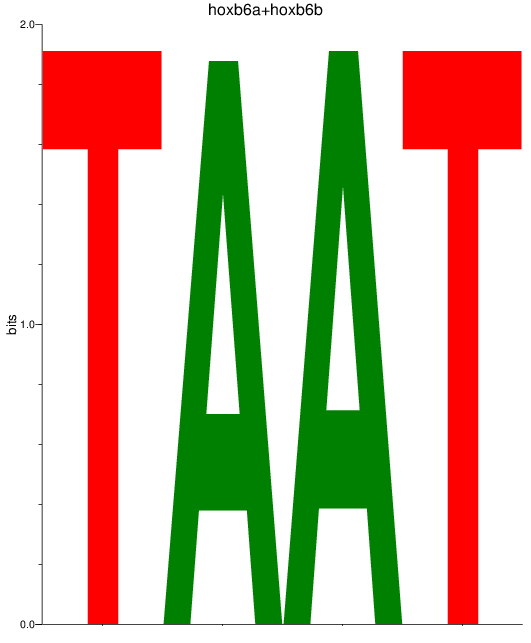
Activity profile of hoxb6a+hoxb6b motif
Sorted Z-values of hoxb6a+hoxb6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30244356 | 11.44 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr1_+_17676745 | 10.52 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr1_+_44439661 | 9.98 |
ENSDART00000100309
|
crybb1l2
|
crystallin, beta B1, like 2 |
| chr25_+_31227747 | 9.44 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr21_+_7582036 | 8.91 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr5_-_29643930 | 8.83 |
ENSDART00000161250
|
grin1b
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1b |
| chr21_-_42007213 | 8.52 |
ENSDART00000188804
ENSDART00000092821 ENSDART00000165743 |
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr5_-_44829719 | 8.36 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr8_-_49431939 | 8.08 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr5_-_30615901 | 8.06 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr5_-_23362602 | 7.85 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr7_+_39446247 | 7.54 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr5_-_30620625 | 7.54 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr19_+_40856534 | 7.44 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr22_+_18389271 | 7.06 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr9_+_34641237 | 6.99 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr19_+_40856807 | 6.78 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr23_-_26535875 | 6.77 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr23_-_6641223 | 6.40 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr10_+_26800213 | 6.37 |
ENSDART00000078996
|
arr3a
|
arrestin 3a, retinal (X-arrestin) |
| chr12_-_25916530 | 6.35 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr15_+_45640906 | 6.22 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr20_-_20821783 | 6.19 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr11_+_25472758 | 6.14 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr20_-_40754794 | 6.14 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr17_+_25414033 | 6.05 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr14_+_35748385 | 6.03 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr3_+_32142382 | 6.01 |
ENSDART00000133035
|
syt5a
|
synaptotagmin Va |
| chr3_-_28120092 | 5.93 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr6_+_40661703 | 5.87 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr15_-_44512461 | 5.85 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr3_-_31079186 | 5.82 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr16_+_37582872 | 5.80 |
ENSDART00000169331
|
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr3_-_41791178 | 5.77 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr11_-_34065718 | 5.70 |
ENSDART00000110608
|
col6a1
|
collagen, type VI, alpha 1 |
| chr5_+_45677781 | 5.69 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr14_+_17376940 | 5.68 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr1_-_43915423 | 5.68 |
ENSDART00000181915
ENSDART00000113673 |
scpp5
|
secretory calcium-binding phosphoprotein 5 |
| chr11_-_11266882 | 5.67 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr23_-_30431333 | 5.62 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr25_+_31267268 | 5.60 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr15_+_19652807 | 5.54 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr17_-_15546862 | 5.53 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr11_+_23993298 | 5.53 |
ENSDART00000186757
ENSDART00000172459 |
chia.1
|
chitinase, acidic.1 |
| chr17_-_16965809 | 5.45 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr4_+_7508316 | 5.45 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr21_-_42007482 | 5.39 |
ENSDART00000075740
|
gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr1_-_14234076 | 5.38 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr2_-_27329667 | 5.35 |
ENSDART00000187490
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr1_+_42225060 | 5.31 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr7_-_45076131 | 5.31 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr25_+_29160102 | 5.29 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr8_+_47633438 | 5.28 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr21_-_22115136 | 5.28 |
ENSDART00000134715
ENSDART00000089246 ENSDART00000139789 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr24_-_7699356 | 5.27 |
ENSDART00000013117
|
syt5b
|
synaptotagmin Vb |
| chr12_+_15002757 | 5.26 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr25_+_19105804 | 5.17 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr20_+_30490682 | 5.15 |
ENSDART00000184871
|
myt1la
|
myelin transcription factor 1-like, a |
| chr14_+_35806605 | 5.12 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr19_-_20270178 | 5.03 |
ENSDART00000144891
ENSDART00000090883 |
gpnmb
|
glycoprotein (transmembrane) nmb |
| chr10_-_20453995 | 4.95 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr23_-_17657348 | 4.94 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr12_+_16440708 | 4.91 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr14_+_49135264 | 4.88 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr13_+_3252950 | 4.86 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr16_-_12173554 | 4.81 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr15_-_12545683 | 4.80 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr14_+_35748206 | 4.77 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr13_+_36764715 | 4.76 |
ENSDART00000111832
ENSDART00000085230 |
atl1
|
atlastin GTPase 1 |
| chr21_+_28958471 | 4.73 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr15_+_6109861 | 4.69 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr12_+_34896956 | 4.66 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr19_+_10339538 | 4.63 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr5_-_41494831 | 4.61 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr13_+_25449681 | 4.59 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr15_-_27710513 | 4.56 |
ENSDART00000005641
ENSDART00000134373 |
lhx1a
|
LIM homeobox 1a |
| chr5_+_26795773 | 4.56 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr5_+_9348284 | 4.54 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr23_-_38497705 | 4.50 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr6_-_27123327 | 4.50 |
ENSDART00000073881
|
agxta
|
alanine-glyoxylate aminotransferase a |
| chr23_+_19590598 | 4.45 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr20_-_40755614 | 4.44 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr20_-_26042070 | 4.40 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr3_-_32170850 | 4.38 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr21_-_13085242 | 4.37 |
ENSDART00000044504
|
zgc:109965
|
zgc:109965 |
| chr21_-_22114625 | 4.37 |
ENSDART00000177426
ENSDART00000135410 |
elmod1
|
ELMO/CED-12 domain containing 1 |
| chr2_-_54387550 | 4.36 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr21_+_10739846 | 4.36 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr23_+_21966447 | 4.34 |
ENSDART00000189378
|
lactbl1a
|
lactamase, beta-like 1a |
| chr3_-_32817274 | 4.33 |
ENSDART00000142582
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr16_+_31802203 | 4.29 |
ENSDART00000058739
ENSDART00000110834 |
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr17_-_12389259 | 4.27 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr24_-_6158933 | 4.24 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr6_+_4872883 | 4.23 |
ENSDART00000186730
ENSDART00000092290 ENSDART00000151674 |
pcdh9
|
protocadherin 9 |
| chr20_-_46554440 | 4.22 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr25_-_32869794 | 4.21 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr14_+_32839535 | 4.17 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr1_-_43905252 | 4.17 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr9_+_31795343 | 4.16 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr22_+_20720808 | 4.15 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr21_-_22737228 | 4.13 |
ENSDART00000151366
|
fbxo40.2
|
F-box protein 40, tandem duplicate 2 |
| chr3_-_32169754 | 4.09 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr7_+_29954709 | 4.07 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr10_+_29698467 | 4.05 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr20_-_38617766 | 4.04 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr18_-_38088099 | 4.04 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr2_+_37227011 | 4.03 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr25_+_19106574 | 4.01 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr10_+_21576909 | 4.00 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr6_-_41229787 | 3.99 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr8_+_19356072 | 3.97 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr5_-_54197084 | 3.87 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr11_-_28911172 | 3.87 |
ENSDART00000168493
|
igsf21a
|
immunoglobin superfamily, member 21a |
| chr3_-_28075756 | 3.87 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr7_-_27685365 | 3.83 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr22_-_20011476 | 3.80 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr4_+_12612723 | 3.77 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr18_+_43365890 | 3.77 |
ENSDART00000173113
|
si:ch211-129p13.1
|
si:ch211-129p13.1 |
| chr11_+_24002503 | 3.76 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr13_+_27314795 | 3.75 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr23_+_17220986 | 3.74 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr19_-_6988837 | 3.73 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr1_-_38815361 | 3.72 |
ENSDART00000148790
ENSDART00000148572 ENSDART00000149080 |
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr18_+_45792035 | 3.71 |
ENSDART00000135045
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr6_+_24817852 | 3.71 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr19_-_10196370 | 3.71 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr2_-_33645411 | 3.70 |
ENSDART00000114663
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr21_-_4032650 | 3.70 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr4_-_16330368 | 3.69 |
ENSDART00000128932
|
epyc
|
epiphycan |
| chr8_+_41533268 | 3.67 |
ENSDART00000142377
|
si:ch211-158d24.2
|
si:ch211-158d24.2 |
| chr23_+_6795531 | 3.65 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr10_-_22845485 | 3.65 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr15_-_34408777 | 3.63 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr12_-_31012741 | 3.62 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7 like 2 |
| chr7_-_49594995 | 3.62 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr4_-_10599062 | 3.57 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr2_+_39021282 | 3.56 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr21_-_27010796 | 3.51 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr16_-_12173399 | 3.50 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr19_-_6873107 | 3.47 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr14_+_16345003 | 3.46 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr14_-_49063157 | 3.45 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr1_+_11977426 | 3.44 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr6_-_50204262 | 3.44 |
ENSDART00000163648
|
raly
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr8_-_14052349 | 3.43 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr1_-_21599219 | 3.43 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr7_+_53541173 | 3.42 |
ENSDART00000159449
|
gramd2aa
|
GRAM domain containing 2Aa |
| chr23_+_20563779 | 3.42 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr7_-_28148310 | 3.41 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr11_+_39672874 | 3.40 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr9_+_18716485 | 3.40 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr21_+_11401247 | 3.39 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr9_+_33357011 | 3.39 |
ENSDART00000088569
|
nyx
|
nyctalopin |
| chr25_+_7494181 | 3.39 |
ENSDART00000165005
|
cat
|
catalase |
| chr7_+_49862837 | 3.39 |
ENSDART00000174315
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr8_-_23081511 | 3.38 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr13_+_3667230 | 3.38 |
ENSDART00000131553
ENSDART00000189841 ENSDART00000183554 ENSDART00000018737 |
qkib
|
QKI, KH domain containing, RNA binding b |
| chr11_+_21050326 | 3.38 |
ENSDART00000065984
|
zgc:113307
|
zgc:113307 |
| chr21_-_39639954 | 3.37 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr21_-_41870029 | 3.36 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr5_+_58372164 | 3.36 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr21_-_21373242 | 3.36 |
ENSDART00000079629
|
ppm1nb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Nb (putative) |
| chr16_-_8280885 | 3.34 |
ENSDART00000129068
|
entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr13_-_33022372 | 3.33 |
ENSDART00000147165
|
rbm25a
|
RNA binding motif protein 25a |
| chr10_-_15128771 | 3.33 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr6_-_38419318 | 3.32 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr25_-_19655820 | 3.32 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr23_+_40460333 | 3.32 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr23_-_19500559 | 3.30 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr7_+_17229282 | 3.30 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr6_+_46341306 | 3.29 |
ENSDART00000111905
|
BX649498.1
|
|
| chr17_+_33226955 | 3.28 |
ENSDART00000063333
|
pomca
|
proopiomelanocortin a |
| chr16_+_46111849 | 3.28 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr3_+_54047342 | 3.27 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr17_-_37156520 | 3.27 |
ENSDART00000145669
|
dtnbb
|
dystrobrevin, beta b |
| chr24_-_24849091 | 3.27 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr22_-_10487490 | 3.26 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr2_-_30770736 | 3.24 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr25_-_9805269 | 3.20 |
ENSDART00000192048
|
lrrc4c
|
leucine rich repeat containing 4C |
| chr10_+_21563986 | 3.20 |
ENSDART00000100600
|
pcdh1a6
|
protocadherin 1 alpha 6 |
| chr15_-_24869826 | 3.20 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr14_+_32838110 | 3.19 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr7_+_17229980 | 3.18 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr14_+_45675306 | 3.18 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr11_+_30817943 | 3.17 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr20_-_25522911 | 3.15 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr3_+_31953145 | 3.15 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr14_-_2199573 | 3.14 |
ENSDART00000124485
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr14_+_45676701 | 3.14 |
ENSDART00000183505
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr21_-_42100471 | 3.14 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr7_-_38638809 | 3.14 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr25_+_27923846 | 3.14 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr9_+_33009284 | 3.13 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr10_+_5689510 | 3.12 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr20_-_29420713 | 3.12 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr22_-_10486477 | 3.11 |
ENSDART00000184366
|
aspn
|
asporin (LRR class 1) |
| chr12_+_24344963 | 3.11 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr1_-_50859053 | 3.11 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr5_+_32222303 | 3.09 |
ENSDART00000051362
|
myhc4
|
myosin heavy chain 4 |
| chr20_-_47732703 | 3.09 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb6a+hoxb6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 19.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 2.8 | 8.4 | GO:0005986 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 2.7 | 10.6 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 2.6 | 10.5 | GO:1990544 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 2.4 | 7.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.9 | 11.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 1.9 | 1.9 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 1.8 | 21.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 1.8 | 7.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.8 | 7.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 1.6 | 4.9 | GO:0021611 | facial nerve formation(GO:0021611) |
| 1.5 | 4.6 | GO:0097378 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 1.5 | 4.6 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 1.3 | 9.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 1.2 | 12.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 1.2 | 4.9 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 1.2 | 10.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 1.1 | 3.3 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 1.1 | 5.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.0 | 5.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.0 | 3.9 | GO:0033605 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 1.0 | 2.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 1.0 | 2.9 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 1.0 | 2.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.9 | 3.6 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.9 | 2.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.9 | 0.9 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.9 | 3.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.8 | 2.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.8 | 9.3 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.8 | 0.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.8 | 5.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.8 | 6.5 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.8 | 4.0 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.8 | 4.8 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.8 | 5.6 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.8 | 8.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.8 | 13.8 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.8 | 6.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.8 | 5.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.7 | 16.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.7 | 3.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.7 | 2.2 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.7 | 2.8 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.7 | 2.8 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.7 | 13.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.7 | 2.8 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.7 | 4.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.7 | 13.2 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.6 | 2.6 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.6 | 2.5 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.6 | 2.5 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.6 | 1.8 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.6 | 2.5 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.6 | 4.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.6 | 1.8 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.6 | 3.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.6 | 7.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.6 | 2.9 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.5 | 2.2 | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.5 | 4.8 | GO:0006833 | water transport(GO:0006833) |
| 0.5 | 2.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 | 14.2 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.5 | 3.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.5 | 2.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.5 | 2.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.5 | 2.0 | GO:0043703 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.5 | 2.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.5 | 1.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.5 | 3.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.5 | 10.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 10.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.5 | 1.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.5 | 2.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.5 | 4.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.5 | 9.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.5 | 1.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.5 | 3.7 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.5 | 1.8 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 1.8 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 2.7 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.4 | 16.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 2.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.4 | 1.3 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.4 | 2.6 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.4 | 3.9 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.4 | 1.7 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.4 | 1.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 0.9 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.4 | 6.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 3.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.4 | 4.1 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.4 | 4.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.4 | 5.3 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 5.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 2.0 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.4 | 1.2 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.4 | 1.6 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 3.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.4 | 1.9 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 13.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.4 | 1.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.4 | 1.5 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.4 | 3.8 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.4 | 13.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.4 | 1.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.4 | 1.9 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.4 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.4 | 1.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.4 | 1.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 8.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.3 | 1.0 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.3 | 7.3 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.3 | 5.6 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.3 | 2.3 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 3.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.3 | 1.3 | GO:0046324 | positive regulation of fat cell differentiation(GO:0045600) regulation of glucose import(GO:0046324) |
| 0.3 | 1.0 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.3 | 1.9 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 0.6 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 5.8 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.3 | 0.6 | GO:0051193 | regulation of glycolytic process(GO:0006110) regulation of nucleotide catabolic process(GO:0030811) regulation of cofactor metabolic process(GO:0051193) regulation of coenzyme metabolic process(GO:0051196) |
| 0.3 | 1.9 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.3 | 2.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.3 | 4.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 1.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.3 | 4.0 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.3 | 1.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 3.4 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.3 | 2.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.3 | 0.9 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.3 | 1.2 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 7.0 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.3 | 0.9 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 60.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.3 | 6.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.3 | 1.5 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.3 | 2.4 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.3 | 0.9 | GO:0007529 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.3 | 1.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 0.9 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 | 1.7 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 6.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 4.5 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.3 | 13.5 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.3 | 1.9 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.3 | 1.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.3 | 1.4 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.3 | 0.8 | GO:0043605 | cellular amide catabolic process(GO:0043605) |
| 0.3 | 0.8 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.3 | 0.8 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 8.7 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.3 | 2.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.3 | 1.8 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.3 | 5.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.3 | 3.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 0.8 | GO:1900158 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.3 | 16.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.3 | 2.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.3 | 1.3 | GO:0030431 | sleep(GO:0030431) |
| 0.3 | 0.8 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 1.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 2.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.9 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 1.2 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 3.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 4.3 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 1.7 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.2 | 22.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.2 | 1.7 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 0.7 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.2 | 1.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 2.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 2.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 2.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 2.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 2.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 0.9 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.2 | 0.4 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 3.0 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.2 | 3.0 | GO:0060038 | cardiac muscle tissue growth(GO:0055017) cardiac muscle cell proliferation(GO:0060038) |
| 0.2 | 0.6 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 1.7 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.2 | 3.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.2 | 2.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 4.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 4.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.1 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.2 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.2 | 2.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 1.7 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.2 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.2 | 4.1 | GO:0097061 | dendritic spine organization(GO:0097061) |
| 0.2 | 2.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 3.9 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.2 | 0.8 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.2 | 0.6 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 3.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 6.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 4.4 | GO:0051588 | regulation of neurotransmitter secretion(GO:0046928) regulation of neurotransmitter transport(GO:0051588) |
| 0.2 | 2.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 6.3 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.2 | 39.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.2 | 2.8 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.2 | 3.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 0.6 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.2 | 1.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.2 | 1.3 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.2 | 1.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 1.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 | 0.7 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 6.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 1.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.2 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 1.4 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.2 | 2.8 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 1.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 2.4 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.2 | 4.9 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 2.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.2 | 2.0 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.2 | 2.7 | GO:0046113 | pyrimidine nucleobase catabolic process(GO:0006208) nucleobase catabolic process(GO:0046113) |
| 0.2 | 2.7 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.2 | 9.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 3.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.2 | 4.8 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.2 | 1.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.2 | 13.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.2 | 1.4 | GO:0072178 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.2 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 1.2 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 4.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 2.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.0 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.3 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 8.2 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 8.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 8.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 1.0 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.4 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 1.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 15.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 1.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 4.4 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 1.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.1 | 2.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 2.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.9 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 2.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 2.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.5 | GO:0033006 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) regulation of mast cell activation involved in immune response(GO:0033006) leukocyte degranulation(GO:0043299) regulation of leukocyte degranulation(GO:0043300) mast cell degranulation(GO:0043303) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 6.8 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.8 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.6 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.4 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 1.0 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 1.7 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 2.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 2.0 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 4.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0046379 | hyaluronan biosynthetic process(GO:0030213) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 10.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 13.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 0.8 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0061687 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) detoxification of inorganic compound(GO:0061687) |
| 0.1 | 2.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 3.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 3.5 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 1.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.6 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.3 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.1 | 0.7 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 1.2 | GO:0006182 | cGMP biosynthetic process(GO:0006182) cGMP metabolic process(GO:0046068) |
| 0.1 | 4.1 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.7 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.7 | GO:1990253 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.1 | 5.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.3 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 4.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.2 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.7 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 3.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.8 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.6 | GO:0097306 | cellular response to alcohol(GO:0097306) |
| 0.1 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 3.6 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.1 | 2.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 2.1 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.8 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 2.1 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 0.5 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 0.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 3.9 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 5.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.4 | GO:1902116 | negative regulation of organelle assembly(GO:1902116) |
| 0.1 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.2 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.7 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 0.5 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.8 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.3 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 4.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 1.1 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.7 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.9 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.6 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 8.8 | GO:0099536 | synaptic signaling(GO:0099536) |
| 0.1 | 2.6 | GO:2001056 | positive regulation of cysteine-type endopeptidase activity(GO:2001056) |
| 0.1 | 0.2 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 0.2 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.2 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.1 | 2.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.7 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.1 | 0.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.1 | 0.1 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.9 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.5 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.8 | GO:0060232 | delamination(GO:0060232) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 2.0 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 2.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 3.6 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.5 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.5 | GO:0014074 | response to purine-containing compound(GO:0014074) response to ATP(GO:0033198) response to organophosphorus(GO:0046683) |
| 0.0 | 4.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.5 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 2.3 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.3 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 2.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.9 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.6 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 1.2 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 3.4 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.7 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 1.4 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 22.6 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.9 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 1.7 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.2 | GO:0051045 | regulation of membrane protein ectodomain proteolysis(GO:0051043) negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.9 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 3.2 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0010952 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 1.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 17.3 | GO:0006811 | ion transport(GO:0006811) |
| 0.0 | 4.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.2 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0048886 | neuromast hair cell differentiation(GO:0048886) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0009266 | response to temperature stimulus(GO:0009266) |
| 0.0 | 1.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.7 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.3 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.5 | 3.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.2 | 12.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 1.2 | 7.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 1.1 | 3.4 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.1 | 37.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.0 | 5.8 | GO:0030891 | VCB complex(GO:0030891) |
| 1.0 | 4.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.9 | 40.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.9 | 16.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.9 | 11.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.9 | 7.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.9 | 3.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.8 | 4.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.8 | 4.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.8 | 34.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.7 | 10.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 2.8 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.6 | 2.5 | GO:0031673 | H zone(GO:0031673) |
| 0.6 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.6 | 4.8 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.6 | 4.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.6 | 7.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.6 | 2.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.5 | 2.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.5 | 10.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.5 | 11.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.4 | 15.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.4 | 1.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 2.7 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.4 | 5.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.4 | 3.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 2.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 1.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.4 | 1.9 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.4 | 2.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.4 | 1.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 23.4 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 2.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 4.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 2.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 6.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.3 | 3.6 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.3 | 2.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 18.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 5.3 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 10.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 2.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.3 | 1.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.3 | 14.3 | GO:0034704 | voltage-gated calcium channel complex(GO:0005891) calcium channel complex(GO:0034704) |
| 0.3 | 1.3 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) dendritic spine membrane(GO:0032591) |
| 0.3 | 3.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.3 | 10.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 5.5 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 5.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 16.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 0.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 4.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 2.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 1.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 9.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.2 | 2.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 4.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 1.6 | GO:0030428 | cell septum(GO:0030428) |
| 0.2 | 0.8 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.2 | 1.9 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.2 | 24.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.2 | 2.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.2 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 11.6 | GO:0030141 | secretory granule(GO:0030141) |
| 0.2 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 4.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 4.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 16.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 3.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 1.5 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 19.8 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 5.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 13.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 3.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 21.4 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 1.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 4.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 3.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 2.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 121.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.2 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 4.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 6.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 2.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 6.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 107.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 7.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.6 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 5.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 10.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.7 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 4.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.3 | GO:0045177 | apical plasma membrane(GO:0016324) apical part of cell(GO:0045177) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 3.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 2.6 | 10.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 2.5 | 7.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 2.4 | 7.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.1 | 32.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.0 | 8.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.0 | 27.7 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 1.9 | 5.8 | GO:0048030 | disaccharide binding(GO:0048030) |
| 1.9 | 5.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.5 | 10.6 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 1.4 | 15.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 1.3 | 3.8 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 1.2 | 4.9 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 1.2 | 6.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 1.2 | 7.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.2 | 4.7 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 1.1 | 2.3 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 1.1 | 3.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 3.1 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 1.0 | 1.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 1.0 | 2.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.9 | 16.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.9 | 2.8 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.9 | 5.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.9 | 3.5 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.8 | 9.3 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.8 | 4.9 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.8 | 7.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.8 | 8.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.8 | 3.9 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.8 | 16.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.8 | 5.3 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.7 | 29.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 2.2 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.7 | 2.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.7 | 4.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.7 | 2.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.7 | 2.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.6 | 3.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 2.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.6 | 2.5 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.6 | 3.0 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.6 | 2.4 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.6 | 4.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.6 | 4.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 7.3 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.6 | 4.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.6 | 2.8 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.6 | 2.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.6 | 4.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.5 | 10.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.5 | 2.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.5 | 2.7 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.5 | 2.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 4.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.5 | 1.5 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.5 | 4.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 1.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.5 | 3.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.5 | 2.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.5 | 6.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.5 | 9.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.5 | 6.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.4 | 2.6 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.4 | 5.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.4 | 5.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.4 | 11.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.4 | 7.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 2.6 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.4 | 3.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.4 | 3.3 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.4 | 24.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.4 | 2.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.4 | 5.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 3.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 1.6 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.4 | 3.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 11.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 1.5 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.4 | 3.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 2.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 1.8 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.4 | 0.7 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) type 1 melanocortin receptor binding(GO:0070996) |
| 0.4 | 1.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 14.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.3 | 3.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.3 | 12.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.3 | 6.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.3 | 4.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.3 | 1.0 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.3 | 1.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.9 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 4.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.3 | 1.2 | GO:0047611 | tubulin deacetylase activity(GO:0042903) acetylspermidine deacetylase activity(GO:0047611) |
| 0.3 | 1.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.3 | 2.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 3.8 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.3 | 1.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.3 | 1.2 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 0.9 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.3 | 0.9 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.3 | 8.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.3 | 1.4 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.3 | 2.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 3.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 3.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 2.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 1.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 1.1 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.3 | 1.4 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.3 | 2.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 1.3 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.8 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.3 | 0.8 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.3 | 1.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 2.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 8.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 2.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 2.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 12.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 1.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 5.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 6.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 2.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.6 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 1.4 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.2 | 1.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 4.8 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 5.0 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 3.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.2 | 8.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 2.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 0.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 3.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.2 | 2.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 2.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 14.9 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.2 | 1.7 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 17.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.6 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.2 | 1.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.6 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.2 | 1.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 2.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 5.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.2 | 3.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.2 | 1.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 9.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 1.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 0.8 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 4.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.8 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 1.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 8.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.5 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 0.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 2.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 2.6 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.2 | 4.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.2 | 3.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 3.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.2 | 1.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.2 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 1.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 1.7 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 3.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 0.5 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.2 | 2.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 2.5 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 1.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 5.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 3.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 3.6 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 1.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.7 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 2.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 2.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.5 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 2.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 6.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 19.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 0.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.6 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 1.6 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.5 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 1.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.2 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 15.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 21.6 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.1 | 4.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 1.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.7 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 6.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.6 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 2.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 1.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.7 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 32.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 3.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 7.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.1 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 1.0 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.1 | 2.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 1.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 1.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 9.4 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.1 | 4.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.6 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 61.7 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 3.2 | GO:0022832 | voltage-gated channel activity(GO:0022832) |
| 0.1 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 8.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.1 | 0.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.1 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 0.3 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 2.8 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.1 | 0.9 | GO:0051371 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.7 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 1.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 1.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.9 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.3 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 1.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 14.0 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 2.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 1.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.6 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.4 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 21.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.7 | 22.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.6 | 5.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.5 | 5.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 1.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 5.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.2 | 1.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.2 | 0.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 3.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 3.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.2 | 7.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 3.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.1 | 3.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 4.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 6.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.8 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 2.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 2.0 | 30.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.2 | 3.6 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.9 | 5.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.8 | 4.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.7 | 1.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.6 | 6.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.5 | 6.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.5 | 1.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.5 | 7.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 4.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 4.1 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.4 | 2.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 1.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 3.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 1.5 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.4 | 9.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.4 | 10.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.4 | 8.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 3.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.3 | 4.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.3 | 5.8 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.3 | 3.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.3 | 5.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 2.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 4.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 3.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 4.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.2 | 2.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 7.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 8.6 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 7.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 2.6 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 1.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 5.8 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 1.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 0.9 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 2.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 2.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.0 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |
| 0.1 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.7 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.1 | 1.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.5 | REACTOME NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Neurotransmitter Release Cycle |
| 0.1 | 5.3 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 2.9 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.1 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 3.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 2.7 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.9 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |