Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
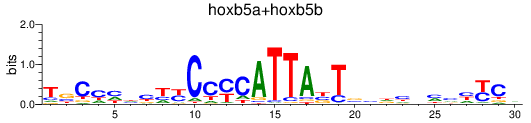
Results for hoxb5a+hoxb5b
Z-value: 0.72
Transcription factors associated with hoxb5a+hoxb5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb5a
|
ENSDARG00000013057 | homeobox B5a |
|
hoxb5b
|
ENSDARG00000054030 | homeobox B5b |
|
hoxb5b
|
ENSDARG00000113174 | homeobox B5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb5a | dr11_v1_chr3_+_23707691_23707691 | -0.82 | 1.7e-05 | Click! |
| hoxb5b | dr11_v1_chr12_+_27129659_27129659 | -0.57 | 1.1e-02 | Click! |
Activity profile of hoxb5a+hoxb5b motif
Sorted Z-values of hoxb5a+hoxb5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_48755638 | 1.78 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr13_+_534453 | 1.57 |
ENSDART00000147909
|
wu:fc17b08
|
wu:fc17b08 |
| chr12_+_7865470 | 1.51 |
ENSDART00000161683
|
BX548028.1
|
|
| chr19_+_17642356 | 1.32 |
ENSDART00000176431
|
CR382334.1
|
|
| chr12_-_7710563 | 1.24 |
ENSDART00000187382
|
CABZ01050062.1
|
|
| chr4_-_77551860 | 1.10 |
ENSDART00000188176
|
AL935186.6
|
|
| chr16_-_29387215 | 1.08 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr4_-_77563411 | 1.06 |
ENSDART00000186841
|
AL935186.8
|
|
| chr21_-_9769500 | 1.05 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr8_-_36554675 | 1.04 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr19_+_15441022 | 0.98 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr6_+_54888493 | 0.94 |
ENSDART00000113331
|
nav1b
|
neuron navigator 1b |
| chr25_+_35212919 | 0.72 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr17_+_53156530 | 0.71 |
ENSDART00000126277
ENSDART00000156774 |
dph6
|
diphthamine biosynthesis 6 |
| chr9_-_12034444 | 0.64 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr6_+_4255319 | 0.63 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr17_+_15041647 | 0.61 |
ENSDART00000108999
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr15_-_29556757 | 0.61 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr5_-_16791223 | 0.56 |
ENSDART00000192744
|
atad1a
|
ATPase family, AAA domain containing 1a |
| chr11_+_24957858 | 0.56 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr13_-_44819254 | 0.53 |
ENSDART00000131673
|
si:dkeyp-2e4.8
|
si:dkeyp-2e4.8 |
| chr3_-_5413018 | 0.53 |
ENSDART00000063138
|
vmo1a
|
vitelline membrane outer layer 1 homolog a |
| chr7_+_56889879 | 0.52 |
ENSDART00000039810
|
myo9aa
|
myosin IXAa |
| chr18_+_33580391 | 0.49 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr14_+_33723309 | 0.48 |
ENSDART00000132488
|
apln
|
apelin |
| chr19_+_48060464 | 0.48 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr4_-_25812329 | 0.47 |
ENSDART00000146658
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr12_+_17436904 | 0.46 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr19_-_27827744 | 0.45 |
ENSDART00000181620
|
papd7
|
PAP associated domain containing 7 |
| chr1_-_56596701 | 0.43 |
ENSDART00000133693
|
LO018605.1
|
|
| chr3_-_4911989 | 0.43 |
ENSDART00000158197
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr22_-_4812755 | 0.42 |
ENSDART00000191739
|
CU570881.2
|
|
| chr1_+_55080797 | 0.42 |
ENSDART00000077390
|
lgalsla
|
lectin, galactoside-binding-like a |
| chr20_+_33708318 | 0.40 |
ENSDART00000135845
|
ak9
|
adenylate kinase 9 |
| chr5_-_71838520 | 0.40 |
ENSDART00000174396
|
CU927890.1
|
|
| chr8_-_43847138 | 0.39 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr15_-_25099679 | 0.38 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr13_-_36134317 | 0.37 |
ENSDART00000015664
ENSDART00000188301 ENSDART00000191073 ENSDART00000184192 ENSDART00000139593 |
pcnx
|
pecanex homolog (Drosophila) |
| chr15_-_684911 | 0.32 |
ENSDART00000161685
|
zgc:171901
|
zgc:171901 |
| chr9_+_6082793 | 0.31 |
ENSDART00000192045
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr6_+_41191482 | 0.31 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr5_-_33769211 | 0.31 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr19_+_46259619 | 0.30 |
ENSDART00000158032
|
grinaa
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1a (glutamate binding) |
| chr25_+_36316280 | 0.30 |
ENSDART00000152735
|
si:ch211-113a14.11
|
si:ch211-113a14.11 |
| chr3_-_1520657 | 0.30 |
ENSDART00000053201
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr14_-_31893996 | 0.30 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr20_+_51730658 | 0.29 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr23_+_22293682 | 0.28 |
ENSDART00000187304
|
CR354612.1
|
|
| chr17_+_49081828 | 0.26 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr5_-_9625459 | 0.26 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr6_-_15603675 | 0.26 |
ENSDART00000143502
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr1_-_59126139 | 0.25 |
ENSDART00000156105
|
si:ch1073-110a20.7
|
si:ch1073-110a20.7 |
| chr11_-_2297832 | 0.25 |
ENSDART00000158266
|
znf740a
|
zinc finger protein 740a |
| chr1_-_51157454 | 0.25 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr18_-_506082 | 0.25 |
ENSDART00000169974
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr3_+_54776117 | 0.24 |
ENSDART00000189047
ENSDART00000115076 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr2_-_3038904 | 0.22 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr6_-_18992896 | 0.22 |
ENSDART00000170228
|
sept9b
|
septin 9b |
| chr15_-_23814330 | 0.22 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr5_+_59392183 | 0.21 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr3_+_54776475 | 0.19 |
ENSDART00000142710
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr10_+_35439952 | 0.19 |
ENSDART00000006284
|
hhla2a.2
|
HERV-H LTR-associating 2a, tandem duplicate 2 |
| chr4_+_41789497 | 0.18 |
ENSDART00000126634
|
si:dkey-237m9.2
|
si:dkey-237m9.2 |
| chr19_-_40175800 | 0.18 |
ENSDART00000102904
|
illr4
|
immune-related, lectin-like receptor 4 |
| chr11_-_6070192 | 0.16 |
ENSDART00000162776
|
babam1
|
BRISC and BRCA1 A complex member 1 |
| chr2_+_24317465 | 0.15 |
ENSDART00000139339
ENSDART00000136284 |
mrpl34
|
mitochondrial ribosomal protein L34 |
| chr7_-_51102479 | 0.15 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr19_+_23298037 | 0.15 |
ENSDART00000183948
|
irgf1
|
immunity-related GTPase family, f1 |
| chr7_+_33314925 | 0.14 |
ENSDART00000148590
|
coro2ba
|
coronin, actin binding protein, 2Ba |
| chr16_+_16826504 | 0.14 |
ENSDART00000108691
|
kcnj14
|
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr10_+_2684958 | 0.14 |
ENSDART00000112019
|
setd9
|
SET domain containing 9 |
| chr10_+_18557102 | 0.14 |
ENSDART00000160157
|
CR388008.1
|
|
| chr4_-_59094319 | 0.12 |
ENSDART00000162560
|
si:dkey-28i19.1
|
si:dkey-28i19.1 |
| chr22_+_17536989 | 0.12 |
ENSDART00000149531
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr5_-_41142467 | 0.11 |
ENSDART00000129415
|
zfr
|
zinc finger RNA binding protein |
| chr13_-_86847 | 0.11 |
ENSDART00000158062
|
pole2
|
polymerase (DNA directed), epsilon 2 |
| chr8_+_36554816 | 0.11 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr10_+_26944418 | 0.09 |
ENSDART00000135493
|
frmd8
|
FERM domain containing 8 |
| chr4_+_77966055 | 0.09 |
ENSDART00000174203
ENSDART00000130100 ENSDART00000080665 ENSDART00000174317 ENSDART00000190123 |
zgc:113921
|
zgc:113921 |
| chr20_+_48543492 | 0.08 |
ENSDART00000175480
|
LO018254.1
|
|
| chr14_-_43586150 | 0.07 |
ENSDART00000113697
|
eif4ea
|
eukaryotic translation initiation factor 4ea |
| chr20_+_43113465 | 0.07 |
ENSDART00000004842
|
dusp23a
|
dual specificity phosphatase 23a |
| chr8_+_25962833 | 0.07 |
ENSDART00000086583
|
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr4_+_38547203 | 0.06 |
ENSDART00000171349
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr1_-_49498116 | 0.06 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr14_+_8476769 | 0.06 |
ENSDART00000131773
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr9_+_16854121 | 0.05 |
ENSDART00000110866
|
cln5
|
CLN5, intracellular trafficking protein |
| chr15_-_35031523 | 0.05 |
ENSDART00000154848
|
si:ch211-272b8.6
|
si:ch211-272b8.6 |
| chr17_+_20569806 | 0.05 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr7_+_36035432 | 0.05 |
ENSDART00000179004
|
irx3a
|
iroquois homeobox 3a |
| chr15_-_41458613 | 0.04 |
ENSDART00000141496
|
nlrc9
|
NLR family CARD domain containing 9 |
| chr2_-_24317240 | 0.04 |
ENSDART00000078975
|
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr23_-_41965557 | 0.04 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr14_-_25078569 | 0.04 |
ENSDART00000172802
ENSDART00000173345 ENSDART00000135004 |
matr3l1.1
|
matrin 3-like 1.1 |
| chr3_+_28831450 | 0.04 |
ENSDART00000055422
|
flr
|
fleer |
| chr7_-_72426484 | 0.04 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr21_-_43117327 | 0.03 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr15_-_2803313 | 0.03 |
ENSDART00000060839
|
tgfb1a
|
transforming growth factor, beta 1a |
| chr9_-_41817712 | 0.02 |
ENSDART00000182657
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr22_+_12477996 | 0.02 |
ENSDART00000177704
|
CR847870.3
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb5a+hoxb5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 1.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.4 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.7 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.5 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 0.4 | GO:0009202 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.5 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.3 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.9 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.2 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.1 | GO:0008016 | regulation of heart contraction(GO:0008016) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 0.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.7 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |