Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
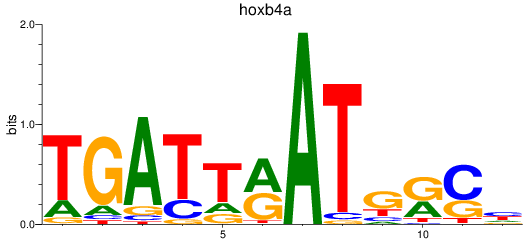
Results for hoxb4a
Z-value: 0.16
Transcription factors associated with hoxb4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb4a
|
ENSDARG00000013533 | homeobox B4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb4a | dr11_v1_chr3_+_23710839_23710839 | 0.17 | 4.8e-01 | Click! |
Activity profile of hoxb4a motif
Sorted Z-values of hoxb4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_29072162 | 0.29 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr19_+_14059349 | 0.28 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr21_+_20901505 | 0.26 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr12_+_28854963 | 0.25 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr12_-_26430507 | 0.23 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr13_+_18321140 | 0.19 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr17_+_27434626 | 0.19 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr21_-_20328375 | 0.18 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr20_-_26491567 | 0.17 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr3_+_27027781 | 0.17 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr17_-_114121 | 0.17 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr12_-_20616160 | 0.15 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr9_-_15789526 | 0.15 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr2_+_44972720 | 0.13 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr3_+_33300522 | 0.12 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr17_-_23241393 | 0.09 |
ENSDART00000190697
|
AL935174.4
|
|
| chr10_-_20445549 | 0.09 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr13_-_33207367 | 0.08 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr5_+_32924669 | 0.08 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr19_+_20177208 | 0.08 |
ENSDART00000166807
|
tra2a
|
transformer 2 alpha homolog |
| chr6_-_57635577 | 0.08 |
ENSDART00000168708
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr23_+_27756984 | 0.07 |
ENSDART00000137103
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr11_-_13341483 | 0.07 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr21_+_22423286 | 0.07 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr9_+_16449398 | 0.07 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr4_+_43408004 | 0.07 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr14_+_46274611 | 0.07 |
ENSDART00000134363
|
cabp2b
|
calcium binding protein 2b |
| chr2_+_27010439 | 0.06 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr6_-_43047774 | 0.06 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr23_+_384850 | 0.06 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr23_+_13814978 | 0.06 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr20_+_51479263 | 0.06 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr19_+_816208 | 0.06 |
ENSDART00000093304
|
nrm
|
nurim |
| chr15_+_25489406 | 0.05 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr11_-_13341051 | 0.05 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr6_-_28345002 | 0.05 |
ENSDART00000158955
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr7_+_13988075 | 0.05 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr7_-_6268373 | 0.04 |
ENSDART00000183990
|
si:ch211-220f21.2
|
si:ch211-220f21.2 |
| chr17_-_8727699 | 0.04 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr16_-_32672883 | 0.04 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr12_+_28854410 | 0.04 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr20_-_32188897 | 0.03 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr1_-_47431453 | 0.03 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr15_-_26552652 | 0.03 |
ENSDART00000152336
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr2_-_54039293 | 0.03 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr17_-_19626357 | 0.03 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr5_-_64103863 | 0.03 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr3_+_23752150 | 0.03 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr18_-_12957451 | 0.03 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr1_+_1712140 | 0.02 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr25_+_5972690 | 0.02 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr2_+_23731194 | 0.02 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr6_-_49655879 | 0.02 |
ENSDART00000191594
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr5_+_20112032 | 0.02 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr13_-_22961605 | 0.02 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr3_-_51109286 | 0.02 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr23_-_3721444 | 0.01 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr7_+_27317174 | 0.01 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_+_28256076 | 0.01 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr3_+_19687217 | 0.01 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr6_+_29791164 | 0.01 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr6_-_29105727 | 0.01 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr7_+_35268880 | 0.01 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr15_+_17251191 | 0.01 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr11_+_18205766 | 0.01 |
ENSDART00000175559
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr15_-_15968883 | 0.01 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr6_+_18367388 | 0.00 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr18_-_25401002 | 0.00 |
ENSDART00000055567
|
gnrhr4
|
gonadotropin releasing hormone receptor 4 |
| chr20_+_51478939 | 0.00 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr4_-_17257435 | 0.00 |
ENSDART00000131973
|
lrmp
|
lymphoid-restricted membrane protein |
| chr13_+_30912385 | 0.00 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.2 | GO:0003071 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |