Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
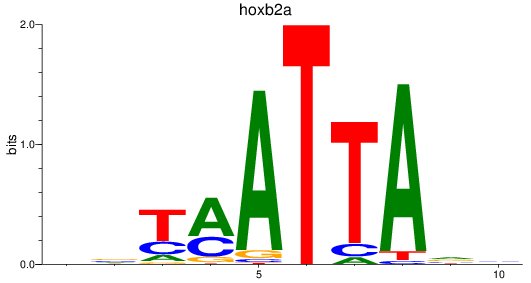
Results for hoxb2a
Z-value: 0.72
Transcription factors associated with hoxb2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb2a
|
ENSDARG00000000175 | homeobox B2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb2a | dr11_v1_chr3_+_23752150_23752150 | 0.56 | 1.3e-02 | Click! |
Activity profile of hoxb2a motif
Sorted Z-values of hoxb2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_14507567 | 4.17 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr21_-_37973819 | 2.12 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr3_-_16719244 | 1.95 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr11_-_35171768 | 1.79 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr1_-_999556 | 1.74 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr8_+_23355484 | 1.70 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr16_+_29509133 | 1.68 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr13_-_21660203 | 1.60 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr18_+_619619 | 1.57 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr10_-_35257458 | 1.44 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr17_-_16422654 | 1.41 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr20_-_27225876 | 1.40 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr1_-_55248496 | 1.40 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr11_-_25418856 | 1.38 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr4_+_9177997 | 1.36 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr8_-_50888806 | 1.36 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr3_+_26244353 | 1.33 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr10_-_44560165 | 1.32 |
ENSDART00000181217
ENSDART00000076084 |
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr17_+_8799451 | 1.30 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_+_30452945 | 1.30 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr12_-_33817114 | 1.30 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr23_-_23401305 | 1.23 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr6_+_41191482 | 1.23 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr17_+_16046132 | 1.20 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr17_+_21295132 | 1.16 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr17_+_16046314 | 1.16 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr15_+_36309070 | 1.15 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr25_-_21031007 | 1.14 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr2_+_33326522 | 1.14 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr16_+_28994709 | 1.12 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr21_+_43404945 | 1.10 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr16_+_5908483 | 1.10 |
ENSDART00000167393
|
ulk4
|
unc-51 like kinase 4 |
| chr21_-_19918286 | 1.08 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr11_+_33818179 | 1.07 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr17_-_31611692 | 1.07 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr19_+_12406583 | 1.06 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr14_-_33945692 | 1.06 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr3_+_22442445 | 1.04 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr21_-_31013817 | 1.04 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr16_+_54209504 | 1.04 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr7_-_25895189 | 1.03 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr18_+_19456648 | 1.02 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr5_+_66433287 | 1.01 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr19_+_31585917 | 1.01 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr21_+_34088110 | 1.01 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr2_-_10600950 | 1.01 |
ENSDART00000160216
|
BX323564.1
|
|
| chr6_-_40922971 | 0.98 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr17_+_24687338 | 0.98 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr10_+_2799285 | 0.97 |
ENSDART00000030709
|
pnx
|
posterior neuron-specific homeobox |
| chr3_-_26244256 | 0.97 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr10_+_22775253 | 0.96 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr10_+_16036246 | 0.96 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr13_+_22295905 | 0.96 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr13_+_35637048 | 0.96 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr13_+_27232848 | 0.95 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr7_+_69019851 | 0.94 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr1_+_513986 | 0.94 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr15_+_1534644 | 0.93 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr3_+_18807006 | 0.92 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr15_-_44052927 | 0.92 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr6_-_43283122 | 0.92 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr10_+_17714866 | 0.92 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr1_+_21731382 | 0.91 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr2_-_23768818 | 0.90 |
ENSDART00000148685
ENSDART00000191167 |
xirp1
|
xin actin binding repeat containing 1 |
| chr15_-_16177603 | 0.89 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr12_-_35830625 | 0.88 |
ENSDART00000180028
|
CU459056.1
|
|
| chr2_+_41526904 | 0.88 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr20_-_23426339 | 0.87 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_-_31695217 | 0.87 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr9_-_32753535 | 0.86 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr22_-_15593824 | 0.85 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr2_-_50272077 | 0.84 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr5_+_27897504 | 0.84 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr8_+_17168114 | 0.83 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr23_+_31912882 | 0.83 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr14_-_25935167 | 0.81 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr25_-_37262220 | 0.79 |
ENSDART00000153789
ENSDART00000155182 |
rfwd3
|
ring finger and WD repeat domain 3 |
| chr24_+_19415124 | 0.78 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr25_-_31423493 | 0.78 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr2_+_20793982 | 0.78 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr23_-_1017605 | 0.76 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr6_+_21001264 | 0.75 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr23_+_31913292 | 0.74 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr12_+_16087077 | 0.74 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr23_+_32028574 | 0.74 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr7_-_17816175 | 0.73 |
ENSDART00000091272
ENSDART00000173757 |
ecsit
|
ECSIT signalling integrator |
| chr15_+_897280 | 0.73 |
ENSDART00000155470
|
znf1012
|
zinc finger protein 1012 |
| chr13_-_42400647 | 0.73 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr15_+_17345609 | 0.71 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr23_-_31913069 | 0.71 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr23_-_20126257 | 0.71 |
ENSDART00000005021
|
tktb
|
transketolase b |
| chr7_+_17543487 | 0.71 |
ENSDART00000126652
|
CU672228.2
|
Danio rerio novel immune-type receptor 1l (nitr1l), mRNA. |
| chr7_+_34290051 | 0.70 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr10_-_41980797 | 0.69 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr17_-_48944465 | 0.69 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr7_+_17106160 | 0.69 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr23_+_4741543 | 0.68 |
ENSDART00000144761
|
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a |
| chr17_+_12658411 | 0.68 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr25_-_27722614 | 0.68 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr4_-_77125693 | 0.68 |
ENSDART00000174256
|
CU467646.3
|
|
| chr3_+_28860283 | 0.67 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr5_-_25733745 | 0.67 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr23_-_31913231 | 0.67 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr5_-_63302944 | 0.67 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr3_-_23643751 | 0.67 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr10_+_7709724 | 0.66 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr5_-_26765188 | 0.66 |
ENSDART00000029450
|
rnf181
|
ring finger protein 181 |
| chr14_+_24845941 | 0.65 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr16_-_28268201 | 0.65 |
ENSDART00000121671
ENSDART00000141911 |
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr4_-_72296520 | 0.64 |
ENSDART00000182638
|
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr19_-_32914227 | 0.64 |
ENSDART00000186115
ENSDART00000124246 |
mtdha
|
metadherin a |
| chr7_+_17816006 | 0.64 |
ENSDART00000080834
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr10_-_21362071 | 0.63 |
ENSDART00000125167
|
avd
|
avidin |
| chr7_+_38936132 | 0.63 |
ENSDART00000173945
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr23_+_44236281 | 0.62 |
ENSDART00000149842
|
MEPCE
|
si:ch1073-157b13.1 |
| chr10_-_21362320 | 0.62 |
ENSDART00000189789
|
avd
|
avidin |
| chr5_+_63302660 | 0.61 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr17_-_22324727 | 0.61 |
ENSDART00000160341
|
CU104709.1
|
|
| chr18_-_20458840 | 0.61 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr22_-_10440688 | 0.61 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr7_+_33132074 | 0.61 |
ENSDART00000073554
|
zgc:153219
|
zgc:153219 |
| chr5_-_14326959 | 0.60 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr2_-_55298075 | 0.60 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr2_+_56213694 | 0.60 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr3_+_30922947 | 0.59 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr8_+_25145464 | 0.59 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr8_+_48484455 | 0.59 |
ENSDART00000122737
|
PRDM16
|
si:ch211-263k4.2 |
| chr8_-_37249813 | 0.59 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr17_+_24722646 | 0.59 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr23_+_11285662 | 0.59 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr18_-_25568994 | 0.58 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr16_-_28878080 | 0.58 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr24_-_4450238 | 0.58 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr3_+_13624815 | 0.58 |
ENSDART00000161451
|
pglyrp6
|
peptidoglycan recognition protein 6 |
| chr22_-_36996856 | 0.58 |
ENSDART00000165923
|
ATP11B
|
si:dkey-211e20.10 |
| chr24_-_34680956 | 0.58 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr20_+_34671386 | 0.57 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr25_+_16116740 | 0.57 |
ENSDART00000139778
|
far1
|
fatty acyl CoA reductase 1 |
| chr17_-_23446760 | 0.57 |
ENSDART00000104715
|
pcgf5a
|
polycomb group ring finger 5a |
| chr5_-_50781623 | 0.57 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr9_-_20372977 | 0.57 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr23_+_44374041 | 0.57 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr9_-_31278048 | 0.57 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr24_-_26981848 | 0.56 |
ENSDART00000183198
|
stag1b
|
stromal antigen 1b |
| chr14_-_33481428 | 0.56 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr12_-_3077395 | 0.56 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_16833518 | 0.56 |
ENSDART00000179867
|
ldhba
|
lactate dehydrogenase Ba |
| chr13_+_30903816 | 0.56 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr19_-_3742472 | 0.56 |
ENSDART00000162132
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr2_+_6253246 | 0.56 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr21_+_34088377 | 0.55 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr8_-_23612462 | 0.55 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr11_-_34219211 | 0.55 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr16_+_9400661 | 0.55 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr17_-_25649079 | 0.55 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr5_+_40837539 | 0.55 |
ENSDART00000188279
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr1_-_26444075 | 0.55 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr20_-_9095105 | 0.54 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr20_-_48898371 | 0.54 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr23_+_42254960 | 0.54 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr1_+_40613297 | 0.54 |
ENSDART00000040798
ENSDART00000168067 ENSDART00000130490 |
naa15b
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit b |
| chr3_+_45365098 | 0.54 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr16_-_17347727 | 0.53 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr25_-_27722309 | 0.53 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr3_+_22035863 | 0.53 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr15_-_34930727 | 0.52 |
ENSDART00000179723
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr22_+_37631234 | 0.52 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr7_+_17816470 | 0.52 |
ENSDART00000173807
|
eml3
|
echinoderm microtubule associated protein like 3 |
| chr10_-_44482911 | 0.52 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr18_-_18875308 | 0.51 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr16_+_16969060 | 0.51 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr5_-_11809710 | 0.51 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr11_+_24703108 | 0.51 |
ENSDART00000159173
|
gpr25
|
G protein-coupled receptor 25 |
| chr24_+_5237753 | 0.51 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr20_+_46513651 | 0.51 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr6_+_40922572 | 0.51 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr19_+_30884706 | 0.51 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr7_+_7511914 | 0.50 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr11_+_31864921 | 0.50 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr6_+_50393047 | 0.50 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr23_-_20345473 | 0.49 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr15_+_7086327 | 0.49 |
ENSDART00000114560
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr21_+_25802190 | 0.49 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr15_+_34988148 | 0.49 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr6_-_35046735 | 0.49 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr21_-_26028205 | 0.49 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr8_+_41037541 | 0.48 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr20_-_38787047 | 0.48 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr4_+_11723852 | 0.48 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr6_+_20647155 | 0.47 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr11_-_30158191 | 0.47 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr7_+_67749251 | 0.47 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr12_-_42214 | 0.47 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr2_+_27010439 | 0.47 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr2_-_59145027 | 0.47 |
ENSDART00000128320
|
FO834803.1
|
|
| chr25_+_35553542 | 0.47 |
ENSDART00000113723
|
spi1a
|
Spi-1 proto-oncogene a |
| chr13_+_36923052 | 0.46 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr18_+_20560616 | 0.46 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr4_+_69559692 | 0.46 |
ENSDART00000164383
|
znf993
|
zinc finger protein 993 |
| chr20_-_54924593 | 0.46 |
ENSDART00000151522
|
si:dkey-15f23.1
|
si:dkey-15f23.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 2.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 1.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.4 | 1.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.4 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 1.7 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.3 | 1.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 1.0 | GO:0061317 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.3 | 1.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.3 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 1.1 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.9 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 0.9 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 3.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 1.0 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.2 | 1.0 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 1.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 0.6 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.2 | 0.6 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.6 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 0.5 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 0.5 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 0.6 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.3 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.4 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 1.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 1.0 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.6 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.5 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.4 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.6 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 1.1 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 1.0 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.7 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.2 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.8 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 1.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 1.6 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.1 | 1.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 4.6 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 0.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.5 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.1 | 0.8 | GO:0051382 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 | 1.0 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.7 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.7 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.1 | 1.7 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.0 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 1.1 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 1.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.7 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.3 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.6 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.2 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.5 | GO:0072595 | maintenance of protein localization in organelle(GO:0072595) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.0 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:0097300 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.3 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 1.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.6 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 1.0 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 2.2 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.4 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.6 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.7 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.5 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.6 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.4 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 1.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 0.6 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 1.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.7 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 1.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 3.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 1.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.9 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 1.7 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 2.0 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 0.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 0.5 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 1.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 1.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.1 | 0.5 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.5 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.6 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.6 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 2.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.9 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.0 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 2.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 1.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.0 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 3.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.8 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 3.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.7 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |