Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
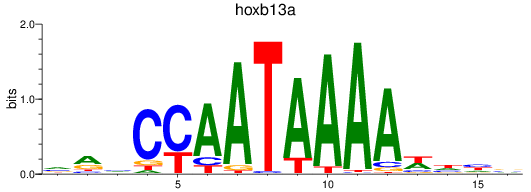
Results for hoxb13a
Z-value: 0.62
Transcription factors associated with hoxb13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb13a
|
ENSDARG00000056015 | homeobox B13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb13a | dr11_v1_chr3_+_23654233_23654233 | 0.50 | 2.7e-02 | Click! |
Activity profile of hoxb13a motif
Sorted Z-values of hoxb13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_15382704 | 2.19 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr12_+_41697664 | 1.92 |
ENSDART00000162302
|
bnip3
|
BCL2 interacting protein 3 |
| chr4_-_4387012 | 1.39 |
ENSDART00000191836
|
CU468826.3
|
Danio rerio U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit-related protein 1-like (LOC100331497), mRNA. |
| chr10_-_14929392 | 1.32 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr9_-_14683574 | 1.31 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr20_-_27733683 | 1.31 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr20_-_25709247 | 1.29 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr1_+_52481332 | 1.25 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr2_+_45548890 | 1.22 |
ENSDART00000113994
|
fndc7a
|
fibronectin type III domain containing 7a |
| chr8_+_25351863 | 1.17 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr5_-_46980651 | 1.13 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr12_-_31457301 | 1.11 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr24_-_23675446 | 1.09 |
ENSDART00000066644
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr10_-_22249444 | 1.06 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr19_-_5380770 | 1.06 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr1_-_9249943 | 1.05 |
ENSDART00000055011
|
zgc:136472
|
zgc:136472 |
| chr25_-_35599887 | 1.04 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr10_+_39164638 | 1.00 |
ENSDART00000188997
|
CU633908.1
|
|
| chr12_+_27704015 | 0.99 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr15_+_45586471 | 0.98 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr19_-_6193448 | 0.97 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr25_-_31739309 | 0.93 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr16_+_21738194 | 0.82 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr20_+_40150612 | 0.81 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr24_-_20808283 | 0.80 |
ENSDART00000143759
|
vipr1b
|
vasoactive intestinal peptide receptor 1b |
| chr25_+_19106574 | 0.77 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr19_-_40776267 | 0.75 |
ENSDART00000189038
|
calcr
|
calcitonin receptor |
| chr23_+_35759843 | 0.69 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr18_+_39327010 | 0.67 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr15_-_28596507 | 0.67 |
ENSDART00000156800
|
si:ch211-225b7.5
|
si:ch211-225b7.5 |
| chr20_-_52338782 | 0.66 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr17_-_7371564 | 0.66 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr11_+_45110865 | 0.65 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr18_-_9046805 | 0.65 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr1_-_1627487 | 0.63 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr25_-_31898552 | 0.63 |
ENSDART00000156128
|
si:ch73-330k17.3
|
si:ch73-330k17.3 |
| chr13_+_1182257 | 0.63 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr5_-_26181863 | 0.62 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr2_-_31661609 | 0.61 |
ENSDART00000144383
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr17_-_52587598 | 0.59 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr18_+_1154189 | 0.59 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr8_-_25120231 | 0.58 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr6_+_49723289 | 0.54 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr20_+_29743904 | 0.53 |
ENSDART00000146366
ENSDART00000153154 |
kidins220b
|
kinase D-interacting substrate 220b |
| chr3_+_46271911 | 0.49 |
ENSDART00000186557
ENSDART00000113531 |
mkl2b
|
MKL/myocardin-like 2b |
| chr11_-_29623380 | 0.48 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr6_+_45300746 | 0.48 |
ENSDART00000159176
|
BX942846.1
|
|
| chr12_-_3705862 | 0.48 |
ENSDART00000193864
ENSDART00000185857 |
CABZ01069040.1
|
|
| chr8_+_10339869 | 0.47 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr22_-_13466246 | 0.45 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr23_+_13124085 | 0.44 |
ENSDART00000139475
|
samd10b
|
sterile alpha motif domain containing 10b |
| chr23_-_7799184 | 0.43 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr24_-_36238054 | 0.41 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr7_-_31781339 | 0.40 |
ENSDART00000142666
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr20_+_34717403 | 0.39 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr2_+_11029138 | 0.38 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr5_-_67750907 | 0.37 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr5_+_22510639 | 0.36 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr16_-_27628994 | 0.36 |
ENSDART00000157407
|
nacad
|
NAC alpha domain containing |
| chr2_+_16597011 | 0.35 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr1_+_2321384 | 0.34 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr25_+_4750972 | 0.32 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr1_-_44940830 | 0.31 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr12_-_34758474 | 0.30 |
ENSDART00000153418
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr22_+_2390544 | 0.30 |
ENSDART00000147345
ENSDART00000133611 |
zgc:171435
|
zgc:171435 |
| chr8_+_30709685 | 0.30 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr2_-_4070850 | 0.29 |
ENSDART00000159990
|
yme1l1b
|
YME1-like 1b |
| chr2_-_31735142 | 0.29 |
ENSDART00000130903
|
ralyl
|
RALY RNA binding protein like |
| chr21_-_37733287 | 0.29 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr1_-_58562129 | 0.28 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr7_-_8324927 | 0.27 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr23_+_30048849 | 0.26 |
ENSDART00000126027
|
uts2a
|
urotensin 2, alpha |
| chr10_+_32066355 | 0.26 |
ENSDART00000062311
|
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr16_+_39485608 | 0.25 |
ENSDART00000188085
|
CR855318.1
|
|
| chr10_+_32066537 | 0.21 |
ENSDART00000124166
|
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr7_+_27251376 | 0.18 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr11_-_30611814 | 0.16 |
ENSDART00000089803
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr24_+_1042594 | 0.16 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr15_+_11381532 | 0.16 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr8_+_25352268 | 0.13 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr17_+_31820401 | 0.11 |
ENSDART00000192607
ENSDART00000157490 |
eef1akmt2
|
EEF1A lysine methyltransferase 2 |
| chr1_-_58561963 | 0.11 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr13_+_17694845 | 0.11 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr6_-_28222592 | 0.11 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr4_-_49987980 | 0.10 |
ENSDART00000150428
|
si:dkey-156k2.4
|
si:dkey-156k2.4 |
| chr9_+_24125445 | 0.07 |
ENSDART00000090346
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr22_+_11857356 | 0.07 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr1_+_47486104 | 0.06 |
ENSDART00000114746
|
lrrc58a
|
leucine rich repeat containing 58a |
| chr11_+_18873619 | 0.06 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr1_+_44941031 | 0.05 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr22_+_18187857 | 0.03 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr19_-_17526735 | 0.03 |
ENSDART00000189391
|
thrb
|
thyroid hormone receptor beta |
| chr7_-_26497947 | 0.02 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr12_-_16524727 | 0.01 |
ENSDART00000166645
|
FO704724.1
|
|
| chr10_-_14929630 | 0.01 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 1.9 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.4 | GO:0031650 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 1.3 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0019482 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.1 | 1.0 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 1.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.8 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.6 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 1.0 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 1.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.8 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.0 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.4 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 1.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 1.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |