Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
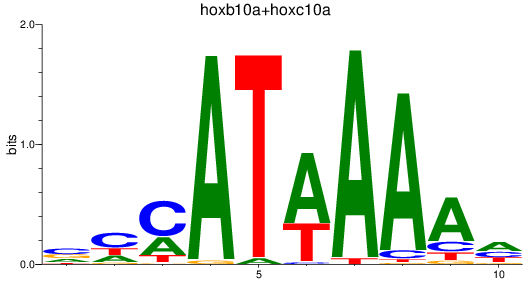
Results for hoxb10a+hoxc10a
Z-value: 0.78
Transcription factors associated with hoxb10a+hoxc10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb10a
|
ENSDARG00000011579 | homeobox B10a |
|
hoxc10a
|
ENSDARG00000070348 | homeobox C10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc10a | dr11_v1_chr23_+_35969228_35969228 | -0.37 | 1.2e-01 | Click! |
| hoxb10a | dr11_v1_chr3_+_23669267_23669267 | 0.05 | 8.4e-01 | Click! |
Activity profile of hoxb10a+hoxc10a motif
Sorted Z-values of hoxb10a+hoxc10a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_2223837 | 1.65 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr14_+_30279391 | 1.49 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr11_+_30244356 | 1.27 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr3_+_54047342 | 1.13 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr4_+_37406676 | 1.11 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr23_+_27068225 | 1.09 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr10_+_26612321 | 0.97 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr23_-_27571667 | 0.94 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr20_+_26538137 | 0.91 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr23_+_33934228 | 0.81 |
ENSDART00000134237
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr1_-_38195012 | 0.80 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr25_+_20089986 | 0.78 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr20_+_9763364 | 0.77 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr24_-_12689571 | 0.71 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr13_+_22264914 | 0.71 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr12_-_13337033 | 0.70 |
ENSDART00000105903
ENSDART00000139786 |
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr7_+_60111581 | 0.68 |
ENSDART00000087093
|
hspa12b
|
heat shock protein 12B |
| chr5_+_23136544 | 0.67 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr10_-_29903165 | 0.65 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr19_-_10881486 | 0.65 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr6_-_60147517 | 0.65 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr6_+_52790049 | 0.64 |
ENSDART00000002571
|
matn4
|
matrilin 4 |
| chr9_-_1939232 | 0.64 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr16_+_50969248 | 0.61 |
ENSDART00000172068
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr16_-_45917322 | 0.60 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr2_-_22659450 | 0.60 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr19_-_22843480 | 0.59 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr15_+_37559570 | 0.59 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr7_-_16562200 | 0.58 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr17_+_33226955 | 0.58 |
ENSDART00000063333
|
pomca
|
proopiomelanocortin a |
| chr2_-_35566938 | 0.57 |
ENSDART00000029006
ENSDART00000077178 ENSDART00000125298 |
tnn
|
tenascin N |
| chr13_+_36585399 | 0.56 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr19_+_5418006 | 0.55 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr8_+_2487883 | 0.55 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr20_+_46183505 | 0.54 |
ENSDART00000060799
|
taar13b
|
trace amine associated receptor 13b |
| chr24_+_1294176 | 0.52 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr7_+_48761875 | 0.52 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr6_-_30485009 | 0.51 |
ENSDART00000025698
|
zgc:153311
|
zgc:153311 |
| chr11_-_6265574 | 0.50 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr19_-_10881141 | 0.49 |
ENSDART00000162793
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr7_-_45076131 | 0.49 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr4_+_7391110 | 0.47 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr16_+_13883872 | 0.47 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr2_-_34072218 | 0.47 |
ENSDART00000184618
|
eif2b3
|
eukaryotic translation initiation factor 2B, subunit 3 gamma |
| chr6_+_35362225 | 0.46 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr7_-_30087048 | 0.46 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr1_-_39983730 | 0.46 |
ENSDART00000160066
|
ing2
|
inhibitor of growth family, member 2 |
| chr6_-_28222592 | 0.45 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr18_+_19974289 | 0.45 |
ENSDART00000090334
ENSDART00000192982 |
skor1b
|
SKI family transcriptional corepressor 1b |
| chr24_-_27473771 | 0.45 |
ENSDART00000139874
|
cxl34b.11
|
CX chemokine ligand 34b, duplicate 11 |
| chr17_+_15535501 | 0.44 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr6_-_37468971 | 0.43 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr24_-_8732519 | 0.42 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr14_-_30967284 | 0.42 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr19_-_47571797 | 0.41 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_+_20396858 | 0.41 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr12_+_27117609 | 0.41 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr5_-_24174956 | 0.40 |
ENSDART00000137631
|
si:ch211-137i24.10
|
si:ch211-137i24.10 |
| chr19_-_47571456 | 0.40 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr17_-_12389259 | 0.40 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr20_-_29499363 | 0.39 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr14_+_23717165 | 0.39 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr9_+_41024973 | 0.38 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr15_+_31344472 | 0.38 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr5_-_23715861 | 0.38 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr17_+_45395846 | 0.37 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr5_+_24245682 | 0.37 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr13_+_35474235 | 0.36 |
ENSDART00000181927
|
mkks
|
McKusick-Kaufman syndrome |
| chr3_-_31804481 | 0.36 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr8_+_3379815 | 0.36 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr10_-_35002731 | 0.35 |
ENSDART00000131279
|
alg5
|
asparagine-linked glycosylation 5 (dolichyl-phosphate beta-glucosyltransferase) |
| chr17_-_15746898 | 0.35 |
ENSDART00000180079
|
cx52.7
|
connexin 52.7 |
| chr21_+_10756154 | 0.35 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr10_+_36650222 | 0.35 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr23_+_43718115 | 0.35 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr18_+_7363242 | 0.35 |
ENSDART00000133375
|
si:dkey-30c15.17
|
si:dkey-30c15.17 |
| chr2_+_35595454 | 0.35 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr3_-_31069776 | 0.34 |
ENSDART00000167462
|
elob
|
elongin B |
| chr16_+_25664387 | 0.34 |
ENSDART00000184394
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr18_+_20825375 | 0.34 |
ENSDART00000090100
ENSDART00000146751 ENSDART00000141686 |
ttc23
|
tetratricopeptide repeat domain 23 |
| chr17_-_15747296 | 0.34 |
ENSDART00000153970
|
cx52.7
|
connexin 52.7 |
| chr17_+_17764979 | 0.34 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr8_+_2487250 | 0.34 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr5_-_54712159 | 0.33 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr6_-_41138854 | 0.33 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr21_+_15733880 | 0.33 |
ENSDART00000149371
ENSDART00000184111 |
idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr10_+_44692272 | 0.33 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr21_-_14826066 | 0.33 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr6_+_13806466 | 0.32 |
ENSDART00000043522
|
tmem198b
|
transmembrane protein 198b |
| chr17_+_33453689 | 0.32 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr6_+_36877968 | 0.32 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr20_+_16173618 | 0.31 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr12_-_20373058 | 0.31 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr21_-_25756119 | 0.30 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr5_-_36597612 | 0.30 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr10_-_25328814 | 0.30 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr4_+_2267641 | 0.30 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr10_+_40606084 | 0.30 |
ENSDART00000133119
|
si:ch211-238p8.24
|
si:ch211-238p8.24 |
| chr9_-_32753535 | 0.30 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr6_+_28208973 | 0.30 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr18_-_15329454 | 0.29 |
ENSDART00000191903
ENSDART00000019818 |
ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr20_-_27390258 | 0.29 |
ENSDART00000010584
|
prph2l
|
peripherin 2, like |
| chr7_-_17457872 | 0.29 |
ENSDART00000191583
ENSDART00000192609 |
nitr3c
|
novel immune-type receptor 3c |
| chr12_+_34051848 | 0.29 |
ENSDART00000153276
|
cyth1b
|
cytohesin 1b |
| chr19_+_19599528 | 0.28 |
ENSDART00000163054
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr6_-_6258451 | 0.28 |
ENSDART00000081966
ENSDART00000125918 |
rtn4a
|
reticulon 4a |
| chr16_+_46459680 | 0.28 |
ENSDART00000101698
|
rpz3
|
rapunzel 3 |
| chr10_+_5234327 | 0.27 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr24_-_27452488 | 0.27 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr12_-_17602958 | 0.27 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr14_+_22114918 | 0.27 |
ENSDART00000166610
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr3_+_39579393 | 0.27 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr5_+_15202495 | 0.27 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr20_-_32045057 | 0.26 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr12_+_19356623 | 0.26 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr19_-_15855427 | 0.25 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr6_-_42036685 | 0.25 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr9_+_38588081 | 0.25 |
ENSDART00000031127
ENSDART00000131784 |
snx4
|
sorting nexin 4 |
| chr11_+_2699951 | 0.25 |
ENSDART00000082512
|
tmem167b
|
transmembrane protein 167B |
| chr7_+_22688781 | 0.24 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr13_-_35984530 | 0.24 |
ENSDART00000143488
|
si:ch211-67f13.8
|
si:ch211-67f13.8 |
| chr6_+_27624023 | 0.24 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr13_-_42749916 | 0.24 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr16_+_25664078 | 0.23 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr10_+_506538 | 0.23 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr4_+_44954486 | 0.23 |
ENSDART00000150875
|
si:dkeyp-100h4.8
|
si:dkeyp-100h4.8 |
| chr10_+_18878856 | 0.23 |
ENSDART00000024127
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr6_-_37749711 | 0.23 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr23_-_43718067 | 0.22 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr8_+_8860902 | 0.22 |
ENSDART00000144986
|
otud5a
|
OTU deubiquitinase 5a |
| chr16_+_7380463 | 0.21 |
ENSDART00000029727
ENSDART00000149086 |
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr20_-_6196989 | 0.21 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr21_-_21594841 | 0.19 |
ENSDART00000132490
|
or133-2
|
odorant receptor, family H, subfamily 133, member 2 |
| chr24_+_12913329 | 0.19 |
ENSDART00000141829
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr20_-_33966148 | 0.19 |
ENSDART00000148111
|
selp
|
selectin P |
| chr16_+_39271123 | 0.19 |
ENSDART00000043823
ENSDART00000141801 |
osbpl10b
|
oxysterol binding protein-like 10b |
| chr7_+_62248514 | 0.19 |
ENSDART00000025308
|
tbc1d19
|
TBC1 domain family, member 19 |
| chr20_+_27087539 | 0.18 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr3_+_46764278 | 0.18 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr15_-_5207805 | 0.18 |
ENSDART00000174046
|
or128-7
|
odorant receptor, family E, subfamily 128, member 7 |
| chr19_+_42061699 | 0.18 |
ENSDART00000125579
|
si:ch211-13c6.2
|
si:ch211-13c6.2 |
| chr23_+_11669337 | 0.18 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr4_-_14191717 | 0.17 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr20_-_33961697 | 0.17 |
ENSDART00000061765
|
selp
|
selectin P |
| chr10_-_34772211 | 0.17 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr16_-_7443388 | 0.17 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr19_+_28187480 | 0.17 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr6_+_13045885 | 0.16 |
ENSDART00000104757
|
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr25_+_4750972 | 0.16 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr12_+_46543572 | 0.16 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr17_-_47090440 | 0.16 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr20_+_46172649 | 0.15 |
ENSDART00000111317
|
taar13e
|
trace amine associated receptor 13e |
| chr5_+_26913120 | 0.15 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr9_-_34937025 | 0.15 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr6_-_31682135 | 0.15 |
ENSDART00000153988
|
cachd1
|
cache domain containing 1 |
| chr2_-_38287987 | 0.15 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr4_-_14191434 | 0.14 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr7_+_58179513 | 0.14 |
ENSDART00000123117
|
ggh
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr14_+_23184517 | 0.14 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr21_-_13225402 | 0.14 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr19_+_19762183 | 0.13 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr22_+_26853254 | 0.13 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr12_-_13155653 | 0.13 |
ENSDART00000152467
|
cxl34c
|
CX chemokine ligand 34c |
| chr2_-_45663945 | 0.13 |
ENSDART00000075080
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr23_+_17417539 | 0.12 |
ENSDART00000182605
|
BX649300.2
|
|
| chr12_+_17603528 | 0.12 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr14_-_31488100 | 0.12 |
ENSDART00000186246
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr5_+_30588856 | 0.12 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr10_-_10969444 | 0.12 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr22_+_18188045 | 0.12 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr11_+_36683859 | 0.11 |
ENSDART00000170102
|
si:ch211-11c3.12
|
si:ch211-11c3.12 |
| chr3_+_32933663 | 0.11 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr10_+_40583930 | 0.11 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr21_-_20328375 | 0.11 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr13_+_45431660 | 0.11 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr8_-_19487327 | 0.11 |
ENSDART00000111710
|
edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr7_-_69025306 | 0.11 |
ENSDART00000180796
|
CABZ01057488.2
|
|
| chr6_+_41191482 | 0.11 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr23_-_9807546 | 0.11 |
ENSDART00000136740
ENSDART00000004474 |
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr21_+_4957685 | 0.10 |
ENSDART00000144166
|
B3GNT10
|
si:ch73-29c22.2 |
| chr7_-_29341233 | 0.10 |
ENSDART00000140938
ENSDART00000147251 |
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr2_+_31948352 | 0.09 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr13_-_26799244 | 0.09 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr4_+_5741733 | 0.09 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr7_-_60096318 | 0.09 |
ENSDART00000189125
|
BX511067.1
|
|
| chr23_+_7379728 | 0.09 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr22_+_1421212 | 0.09 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr22_+_1779401 | 0.08 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr3_+_35812040 | 0.08 |
ENSDART00000075903
ENSDART00000147712 |
crlf3
|
cytokine receptor-like factor 3 |
| chr2_-_28420415 | 0.08 |
ENSDART00000183857
|
CABZ01056051.1
|
|
| chr2_+_24374305 | 0.08 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr24_-_27461295 | 0.07 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr5_-_48307804 | 0.07 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr13_-_50546634 | 0.07 |
ENSDART00000192127
|
CU570781.2
|
|
| chr8_+_30709685 | 0.07 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr6_+_13083146 | 0.07 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr10_+_11265387 | 0.07 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr19_-_17385548 | 0.07 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr11_-_18253111 | 0.07 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr20_-_31497300 | 0.06 |
ENSDART00000046841
|
sash1a
|
SAM and SH3 domain containing 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb10a+hoxc10a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.9 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.6 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.8 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.1 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 1.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.4 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.3 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 1.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.9 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.5 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.5 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.1 | 0.3 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.4 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 0.3 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 0.2 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.0 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.4 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 1.0 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 1.3 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 1.2 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 1.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.3 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.6 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:0071715 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.2 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.5 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 1.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.9 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) translation preinitiation complex(GO:0070993) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 1.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.6 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.4 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 1.0 | GO:0015185 | gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.5 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.1 | 0.8 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.4 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 1.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |