Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
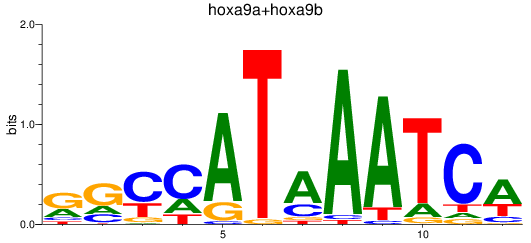
Results for hoxa9a+hoxa9b
Z-value: 1.74
Transcription factors associated with hoxa9a+hoxa9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa9b
|
ENSDARG00000056819 | homeobox A9b |
|
hoxa9a
|
ENSDARG00000105013 | homeobox A9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa9b | dr11_v1_chr16_+_20915319_20915319 | -0.96 | 7.5e-11 | Click! |
| hoxa9a | dr11_v1_chr19_+_19750101_19750223 | -0.96 | 1.3e-10 | Click! |
Activity profile of hoxa9a+hoxa9b motif
Sorted Z-values of hoxa9a+hoxa9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_46875310 | 10.72 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr8_+_6954984 | 9.59 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr7_+_39386982 | 9.08 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr5_+_2815021 | 6.53 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr10_-_11385155 | 6.31 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr20_+_6142433 | 6.25 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr12_-_20362041 | 5.68 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr2_-_3437862 | 5.62 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr8_+_47633438 | 5.49 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr9_-_22299412 | 5.15 |
ENSDART00000139101
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr19_-_31042570 | 4.99 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_32421046 | 4.89 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr6_+_41186320 | 4.75 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr6_+_27151940 | 4.70 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr24_-_40009446 | 4.69 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr24_-_4765740 | 4.65 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr2_+_2223837 | 4.63 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr6_+_7444899 | 4.27 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr14_+_45675306 | 4.24 |
ENSDART00000105461
|
rom1b
|
retinal outer segment membrane protein 1b |
| chr21_+_11401247 | 4.16 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr14_-_50892442 | 4.04 |
ENSDART00000174562
|
CDHR2
|
cadherin related family member 2 |
| chr21_+_11969603 | 3.89 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr11_-_11266882 | 3.69 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr9_-_9992697 | 3.62 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr11_-_6188413 | 3.48 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr16_-_17175731 | 3.48 |
ENSDART00000183057
|
opn9
|
opsin 9 |
| chr9_-_12575776 | 3.41 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr25_-_30429607 | 3.37 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr8_-_39739627 | 3.31 |
ENSDART00000135422
ENSDART00000067844 |
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr4_+_10017049 | 3.29 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr13_+_19884631 | 3.06 |
ENSDART00000089533
|
atrnl1a
|
attractin-like 1a |
| chr13_-_290377 | 3.00 |
ENSDART00000134963
|
chs1
|
chitin synthase 1 |
| chr11_-_141592 | 2.95 |
ENSDART00000092787
|
cdk4
|
cyclin-dependent kinase 4 |
| chr13_+_25380432 | 2.93 |
ENSDART00000038524
|
gsto1
|
glutathione S-transferase omega 1 |
| chr25_-_7999756 | 2.89 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr20_-_29418620 | 2.86 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr3_+_32526263 | 2.83 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr20_-_25533739 | 2.79 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr21_-_26089964 | 2.76 |
ENSDART00000027848
|
tlcd1
|
TLC domain containing 1 |
| chr7_+_48761875 | 2.76 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr13_+_52061034 | 2.76 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr5_+_32206378 | 2.75 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr20_-_20821783 | 2.73 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr8_-_53198154 | 2.63 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr9_+_13714379 | 2.63 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr5_+_9382301 | 2.60 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr13_-_44285793 | 2.56 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr4_+_7841627 | 2.55 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr3_+_34821327 | 2.49 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr3_+_31953145 | 2.44 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr4_+_72797711 | 2.39 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr12_+_16440708 | 2.39 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr20_+_29587995 | 2.38 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr10_+_24627683 | 2.37 |
ENSDART00000112652
|
slc46a3
|
solute carrier family 46, member 3 |
| chr5_+_9377005 | 2.36 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr3_+_4997545 | 2.34 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr18_+_21122818 | 2.29 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr7_-_38634845 | 2.27 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr2_+_16846772 | 2.27 |
ENSDART00000183564
ENSDART00000126718 |
fam131a
|
family with sequence similarity 131, member A |
| chr8_+_14792830 | 2.27 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr24_-_11325849 | 2.23 |
ENSDART00000182485
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr8_+_26859639 | 2.22 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr5_+_36932718 | 2.20 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr3_+_1223824 | 2.16 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr16_+_50289916 | 2.12 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr9_-_6380653 | 2.11 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr19_-_6385594 | 2.07 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr15_+_8043751 | 2.06 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr1_-_52201266 | 2.06 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr3_-_11922713 | 2.04 |
ENSDART00000150575
|
coro7
|
coronin 7 |
| chr19_+_32321797 | 2.04 |
ENSDART00000167664
|
atxn1a
|
ataxin 1a |
| chr22_+_7480465 | 2.03 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr16_+_46111849 | 2.03 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr14_-_31893996 | 2.01 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr7_-_24047316 | 1.97 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr20_-_32110882 | 1.96 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr8_+_32402441 | 1.96 |
ENSDART00000191451
|
epgn
|
epithelial mitogen homolog (mouse) |
| chr3_-_48716422 | 1.96 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr21_+_39100289 | 1.92 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr17_-_28749640 | 1.92 |
ENSDART00000000948
|
coch
|
coagulation factor C homolog, cochlin (Limulus polyphemus) |
| chr11_+_42730639 | 1.88 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr4_-_27301356 | 1.87 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr11_+_6159595 | 1.85 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr9_-_2572790 | 1.84 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr12_-_26406323 | 1.83 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr21_-_25685739 | 1.81 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr9_-_12575569 | 1.80 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr1_+_54677173 | 1.79 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr16_+_3982590 | 1.78 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr17_-_6399920 | 1.78 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr7_-_20731078 | 1.76 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr6_-_51101834 | 1.76 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr7_-_4036184 | 1.76 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr23_+_39963599 | 1.75 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr3_-_24980067 | 1.75 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr6_-_24087279 | 1.75 |
ENSDART00000163445
|
zgc:109982
|
zgc:109982 |
| chr7_-_41693004 | 1.72 |
ENSDART00000121509
|
malrd1
|
MAM and LDL receptor class A domain containing 1 |
| chr11_-_9808356 | 1.72 |
ENSDART00000167465
|
nlgn1
|
neuroligin 1 |
| chr16_-_27749172 | 1.71 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr10_-_19801821 | 1.71 |
ENSDART00000148013
|
gfra2b
|
GDNF family receptor alpha 2b |
| chr16_+_12836143 | 1.70 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr14_-_2036604 | 1.69 |
ENSDART00000192446
|
BX005294.2
|
|
| chr16_-_46645396 | 1.69 |
ENSDART00000131485
|
tmem176l.2
|
transmembrane protein 176l.2 |
| chr19_-_10425140 | 1.68 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr25_-_3393705 | 1.68 |
ENSDART00000163096
|
gpr22b
|
G protein-coupled receptor 22b |
| chr10_+_37145007 | 1.68 |
ENSDART00000131777
|
cuedc1a
|
CUE domain containing 1a |
| chr2_+_927204 | 1.64 |
ENSDART00000165477
|
AL935300.3
|
|
| chr9_-_2573121 | 1.64 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr10_+_439692 | 1.58 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr7_-_4461104 | 1.57 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr18_+_13792490 | 1.56 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr4_+_21129752 | 1.55 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr20_-_34754617 | 1.54 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr1_-_46401385 | 1.53 |
ENSDART00000150029
|
atp11a
|
ATPase phospholipid transporting 11A |
| chr7_-_24046999 | 1.53 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr20_+_52595220 | 1.52 |
ENSDART00000180610
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr7_+_69841017 | 1.50 |
ENSDART00000169107
|
FO818704.1
|
|
| chr5_-_71838520 | 1.49 |
ENSDART00000174396
|
CU927890.1
|
|
| chr16_-_17188294 | 1.49 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr18_+_28102620 | 1.48 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr17_-_51262430 | 1.48 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr6_-_44711942 | 1.48 |
ENSDART00000055035
|
cntn3b
|
contactin 3b |
| chr2_+_6926100 | 1.48 |
ENSDART00000153289
|
nos1apb
|
nitric oxide synthase 1 (neuronal) adaptor protein b |
| chr11_-_44030962 | 1.47 |
ENSDART00000171910
|
FP016005.1
|
|
| chr13_+_844150 | 1.46 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr5_+_9408901 | 1.46 |
ENSDART00000193364
|
FP236810.1
|
|
| chr23_+_3721042 | 1.45 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr7_+_48761646 | 1.44 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr3_-_3209432 | 1.43 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr10_+_21722892 | 1.42 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr11_+_44356504 | 1.42 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr22_-_38480186 | 1.41 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr8_+_6576940 | 1.41 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr5_-_62940851 | 1.41 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr1_+_7546259 | 1.40 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr7_-_29021757 | 1.39 |
ENSDART00000086905
|
nrn1lb
|
neuritin 1-like b |
| chr6_+_55032439 | 1.37 |
ENSDART00000164232
ENSDART00000158845 ENSDART00000157584 ENSDART00000026359 ENSDART00000122794 ENSDART00000183742 |
mybphb
|
myosin binding protein Hb |
| chr20_-_16548912 | 1.37 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr7_-_41964877 | 1.35 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr9_-_296169 | 1.33 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr19_-_36234185 | 1.33 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr2_-_8609653 | 1.33 |
ENSDART00000193354
ENSDART00000189489 ENSDART00000186144 |
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr25_-_13842618 | 1.32 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr12_+_18681477 | 1.32 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr17_+_50701748 | 1.32 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr1_+_32528097 | 1.31 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr13_-_50002852 | 1.31 |
ENSDART00000099439
|
lyst
|
lysosomal trafficking regulator |
| chr10_-_35410518 | 1.31 |
ENSDART00000048430
|
gabrr3a
|
gamma-aminobutyric acid (GABA) A receptor, rho 3a |
| chr20_-_8365244 | 1.30 |
ENSDART00000167102
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr22_-_11493236 | 1.30 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr2_-_10338759 | 1.29 |
ENSDART00000150166
ENSDART00000149584 |
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr16_+_12281032 | 1.28 |
ENSDART00000138638
|
si:dkey-26c10.5
|
si:dkey-26c10.5 |
| chr9_+_29589790 | 1.28 |
ENSDART00000140388
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr23_+_11669337 | 1.27 |
ENSDART00000131355
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr14_-_2221877 | 1.27 |
ENSDART00000106704
|
pcdh2ab1
|
protocadherin 2 alpha b 1 |
| chr5_-_32274383 | 1.27 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr12_-_4540564 | 1.27 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr12_+_26467847 | 1.27 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr1_+_52929185 | 1.26 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr17_-_52587598 | 1.26 |
ENSDART00000061497
|
si:ch211-173a9.6
|
si:ch211-173a9.6 |
| chr21_-_7882905 | 1.25 |
ENSDART00000056561
|
s100z
|
S100 calcium binding protein Z |
| chr7_+_38529263 | 1.25 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr7_-_69853453 | 1.25 |
ENSDART00000049928
|
myoz2a
|
myozenin 2a |
| chr25_-_35497055 | 1.25 |
ENSDART00000009271
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr18_-_42172101 | 1.24 |
ENSDART00000124211
|
cntn5
|
contactin 5 |
| chr10_-_8053753 | 1.24 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr24_-_21404367 | 1.24 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr7_+_31879986 | 1.23 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr11_-_6048490 | 1.23 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr13_+_12120664 | 1.22 |
ENSDART00000130007
ENSDART00000079443 |
gabra2a
|
gamma-aminobutyric acid type A receptor alpha2 subunit a |
| chr2_+_29884363 | 1.22 |
ENSDART00000139247
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr4_+_22480169 | 1.21 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr10_+_26571174 | 1.20 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr20_+_54037138 | 1.19 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr11_+_36243774 | 1.18 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr12_+_27213733 | 1.18 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr8_-_34065573 | 1.18 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr11_+_44356707 | 1.16 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr18_+_39649660 | 1.16 |
ENSDART00000149859
|
gldn
|
gliomedin |
| chr20_+_33532296 | 1.16 |
ENSDART00000153153
|
kcnf1a
|
potassium voltage-gated channel, subfamily F, member 1a |
| chr23_+_40951443 | 1.16 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr11_-_37997419 | 1.15 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr24_-_23998897 | 1.15 |
ENSDART00000130053
|
zmp:0000000991
|
zmp:0000000991 |
| chr9_+_54686686 | 1.15 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr15_-_1843831 | 1.13 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr24_-_29586082 | 1.12 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr17_-_465285 | 1.12 |
ENSDART00000168718
|
chrm5a
|
cholinergic receptor, muscarinic 5a |
| chr2_-_48375342 | 1.11 |
ENSDART00000148788
|
per2
|
period circadian clock 2 |
| chr2_+_38554260 | 1.11 |
ENSDART00000171527
|
cdh24b
|
cadherin 24, type 2b |
| chr10_+_44924684 | 1.11 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
| chr21_+_22158744 | 1.11 |
ENSDART00000101732
|
capn5b
|
calpain 5b |
| chr2_-_43191465 | 1.11 |
ENSDART00000025254
|
crema
|
cAMP responsive element modulator a |
| chr1_-_23157583 | 1.11 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr11_-_22997506 | 1.09 |
ENSDART00000167817
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr14_-_11529311 | 1.09 |
ENSDART00000127208
|
si:ch211-153b23.7
|
si:ch211-153b23.7 |
| chr15_-_28085480 | 1.08 |
ENSDART00000060304
ENSDART00000192239 |
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr13_-_37620091 | 1.08 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr15_-_34845414 | 1.07 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr22_-_31517300 | 1.07 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr9_+_21820101 | 1.06 |
ENSDART00000145431
|
txndc9
|
thioredoxin domain containing 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa9a+hoxa9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.9 | 4.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.7 | 2.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.7 | 4.2 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.7 | 2.8 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.6 | 6.3 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.6 | 3.0 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.5 | 5.9 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.5 | 3.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.5 | 2.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.5 | 4.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 1.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 1.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.4 | 1.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.4 | 2.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.4 | 1.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 1.7 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.3 | 1.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.3 | 2.1 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.3 | 0.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.3 | 3.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 4.6 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 0.7 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.2 | 0.9 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.2 | 0.7 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.2 | 1.7 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 2.0 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 1.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 2.7 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.6 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.8 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 1.4 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.9 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 1.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.2 | 9.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.2 | 1.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.2 | 9.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 0.7 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.8 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 1.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 2.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 0.9 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 2.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 2.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 1.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 1.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.6 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 1.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 3.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.8 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 3.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 1.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 2.0 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 1.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 3.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 7.9 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.8 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.5 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.1 | GO:0097623 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.1 | 2.3 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.1 | 5.3 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.3 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.1 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 6.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 7.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 13.3 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.8 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.2 | GO:2000425 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 0.1 | 0.3 | GO:0032965 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.1 | 1.0 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 2.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 1.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 1.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.3 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 2.9 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.0 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0071938 | vitamin transmembrane transport(GO:0035461) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.8 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 3.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.3 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.1 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 3.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.8 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 1.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 2.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 1.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.7 | GO:0001763 | morphogenesis of a branching structure(GO:0001763) |
| 0.0 | 1.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 3.5 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 4.1 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 2.0 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.3 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 1.5 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 3.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.8 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.9 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.8 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 1.1 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.5 | 2.5 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 2.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 2.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 3.0 | GO:0030428 | cell septum(GO:0030428) |
| 0.3 | 1.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 1.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 1.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.2 | 1.7 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 9.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 0.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 5.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 2.4 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 5.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 2.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 2.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.0 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 3.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 4.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 5.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.9 | GO:0031256 | leading edge membrane(GO:0031256) |
| 0.0 | 1.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 4.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 2.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.0 | 2.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 2.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 2.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.5 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 20.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.8 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.5 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 1.9 | 5.6 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 1.6 | 6.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.5 | 5.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.4 | 4.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 2.9 | GO:0045174 | glutathione dehydrogenase (ascorbate) activity(GO:0045174) |
| 0.9 | 4.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.7 | 3.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.6 | 5.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.6 | 2.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.6 | 2.9 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 2.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 3.7 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.5 | 2.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 2.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 1.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 3.0 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.3 | 4.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 1.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.3 | 3.1 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 2.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 2.0 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 1.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.2 | 0.7 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.2 | 0.7 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.2 | 0.7 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.2 | 2.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 0.6 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.2 | 2.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 9.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 2.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.9 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 0.7 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.2 | 6.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 1.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 0.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.2 | 1.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.2 | 1.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 9.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.8 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.2 | 0.6 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 5.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.8 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 1.1 | GO:0016917 | G-protein coupled GABA receptor activity(GO:0004965) GABA receptor activity(GO:0016917) |
| 0.1 | 1.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 1.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.6 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.7 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 2.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 2.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.2 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.1 | 1.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.6 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.9 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 5.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.8 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 5.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 2.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.6 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.1 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 1.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 4.3 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.8 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 5.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 1.4 | GO:0020037 | heme binding(GO:0020037) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 7.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.8 | 4.8 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 0.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.3 | 6.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.3 | 1.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 0.6 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.6 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |