Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
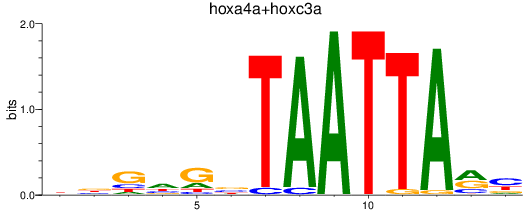
Results for hoxa4a+hoxc3a
Z-value: 0.89
Transcription factors associated with hoxa4a+hoxc3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc3a
|
ENSDARG00000070339 | homeobox C3a |
|
hoxa4a
|
ENSDARG00000103862 | homeobox A4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa3a | dr11_v1_chr19_+_19775757_19775757 | -0.70 | 8.8e-04 | Click! |
| hoxc3a | dr11_v1_chr23_+_36106790_36106790 | -0.68 | 1.4e-03 | Click! |
Activity profile of hoxa4a+hoxc3a motif
Sorted Z-values of hoxa4a+hoxc3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22099536 | 4.50 |
ENSDART00000101923
|
CR391987.1
|
|
| chr11_+_30244356 | 4.46 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr9_-_22129788 | 4.17 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr14_+_46313396 | 3.85 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr9_-_22310919 | 3.37 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr3_-_50443607 | 3.09 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr25_+_29160102 | 2.59 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr2_+_50608099 | 2.54 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr17_-_37395460 | 2.52 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr5_+_32206378 | 2.36 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr12_-_464007 | 2.15 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr16_+_31804590 | 2.06 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr22_+_7738966 | 1.82 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr3_+_32553714 | 1.77 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr15_-_12011390 | 1.71 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr2_+_11205795 | 1.69 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr5_-_71705191 | 1.56 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr20_-_9462433 | 1.51 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr22_-_20011476 | 1.50 |
ENSDART00000093312
ENSDART00000093310 |
celf5a
|
cugbp, Elav-like family member 5a |
| chr9_+_31795343 | 1.50 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr18_+_924949 | 1.49 |
ENSDART00000170888
ENSDART00000193163 |
pkma
|
pyruvate kinase M1/2a |
| chr4_+_9669717 | 1.44 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr3_-_31079186 | 1.42 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr7_+_35075847 | 1.42 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr11_+_3254524 | 1.41 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr7_+_17229980 | 1.39 |
ENSDART00000184910
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr19_+_43297546 | 1.33 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr4_+_21129752 | 1.30 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr19_+_19976990 | 1.27 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr14_+_22172047 | 1.25 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr13_+_33117528 | 1.24 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr12_-_35787801 | 1.23 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr1_-_23308225 | 1.22 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr10_-_27049170 | 1.20 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr1_+_55755304 | 1.16 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr21_+_23108420 | 1.16 |
ENSDART00000192394
ENSDART00000088459 |
htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B |
| chr10_-_5847655 | 1.15 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr21_+_28958471 | 1.14 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr1_-_50859053 | 1.14 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr24_+_16547035 | 1.14 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr7_+_31891110 | 1.10 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr18_+_39487486 | 1.09 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr5_-_51619742 | 1.07 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr15_-_46779934 | 1.07 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr10_-_26744131 | 1.06 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr7_-_28148310 | 1.06 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr21_-_37790727 | 1.06 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr23_+_45584223 | 1.06 |
ENSDART00000149367
|
si:ch73-290k24.5
|
si:ch73-290k24.5 |
| chr16_+_17389116 | 1.04 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr7_+_13418812 | 1.03 |
ENSDART00000191905
ENSDART00000091567 |
dagla
|
diacylglycerol lipase, alpha |
| chr3_+_34919810 | 1.00 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr20_+_4060839 | 0.98 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr12_-_314899 | 0.97 |
ENSDART00000066579
|
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr10_+_15255198 | 0.97 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr20_-_14925281 | 0.97 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr4_+_74929427 | 0.96 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr25_+_3326885 | 0.96 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr7_+_25033924 | 0.96 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr19_-_5103141 | 0.95 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr7_+_30787903 | 0.94 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr24_+_13316737 | 0.94 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr13_-_29420885 | 0.91 |
ENSDART00000024225
|
chata
|
choline O-acetyltransferase a |
| chr2_+_5371492 | 0.89 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr7_+_17229282 | 0.89 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr23_-_20051369 | 0.89 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr16_+_13818500 | 0.89 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr12_-_33357655 | 0.88 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr10_+_15255012 | 0.88 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr16_-_13818061 | 0.88 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr2_-_14390627 | 0.87 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr23_+_20110086 | 0.87 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr25_-_13842618 | 0.87 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr3_-_20040636 | 0.86 |
ENSDART00000104118
|
atxn7l3
|
ataxin 7-like 3 |
| chr14_-_30587814 | 0.85 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr12_+_20352400 | 0.85 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr10_+_16188761 | 0.85 |
ENSDART00000193244
|
ctxn3
|
cortexin 3 |
| chr3_+_23092762 | 0.83 |
ENSDART00000142884
ENSDART00000024136 |
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr3_+_17537352 | 0.83 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr16_-_28658341 | 0.81 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr22_+_28337429 | 0.80 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr9_-_35334642 | 0.80 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr6_+_9870192 | 0.80 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr18_+_8901846 | 0.80 |
ENSDART00000132109
ENSDART00000144247 |
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr5_-_50992690 | 0.80 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr22_-_10459880 | 0.79 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr19_-_5103313 | 0.79 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr23_+_40460333 | 0.78 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr5_+_37068223 | 0.75 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr8_-_23780334 | 0.74 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr23_+_28322986 | 0.74 |
ENSDART00000134710
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr14_-_858985 | 0.73 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr12_-_15584479 | 0.72 |
ENSDART00000150134
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr21_-_14251306 | 0.72 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr12_+_22580579 | 0.72 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr5_+_56023186 | 0.71 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr11_+_44356707 | 0.71 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr16_-_50203058 | 0.71 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr21_-_16113799 | 0.70 |
ENSDART00000187848
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr10_-_31015535 | 0.70 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr4_+_12612723 | 0.68 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr9_-_33725972 | 0.68 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr20_+_591505 | 0.67 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr19_-_27858033 | 0.66 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr21_-_43015383 | 0.66 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr22_-_21476836 | 0.65 |
ENSDART00000135977
|
s1pr3a
|
sphingosine-1-phosphate receptor 3a |
| chr11_+_30057762 | 0.64 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr2_-_28671139 | 0.64 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr16_-_11798994 | 0.64 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr22_-_10121880 | 0.63 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_-_15324837 | 0.63 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr18_+_27571448 | 0.62 |
ENSDART00000147886
|
cd82b
|
CD82 molecule b |
| chr3_+_62353650 | 0.62 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr22_-_26865181 | 0.61 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr20_+_27020201 | 0.61 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr14_-_33936524 | 0.61 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr21_-_32301109 | 0.61 |
ENSDART00000139890
|
clk4b
|
CDC-like kinase 4b |
| chr7_+_34620418 | 0.60 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr16_-_25568512 | 0.60 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr20_+_38032143 | 0.60 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr25_+_3327071 | 0.59 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr10_-_15963903 | 0.59 |
ENSDART00000142357
|
si:dkey-3h23.3
|
si:dkey-3h23.3 |
| chr5_-_4418555 | 0.59 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr6_+_28208973 | 0.59 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr22_-_38621438 | 0.58 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr20_+_37294112 | 0.58 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr7_-_33960170 | 0.58 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr18_-_48547564 | 0.58 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr20_+_19512727 | 0.57 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr20_-_28800999 | 0.57 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr16_-_20312146 | 0.56 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr20_-_14924858 | 0.56 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr21_-_5799122 | 0.56 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr13_+_7292061 | 0.56 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr13_+_4225173 | 0.56 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr13_+_9432501 | 0.55 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr8_-_31384607 | 0.55 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr11_+_44356504 | 0.54 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr9_+_29643036 | 0.53 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr3_-_15080226 | 0.53 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr14_-_30474346 | 0.52 |
ENSDART00000173051
ENSDART00000173124 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr22_+_14051894 | 0.52 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr9_-_43538328 | 0.52 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr12_-_41684729 | 0.51 |
ENSDART00000184461
|
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr21_-_32060993 | 0.51 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr13_-_2010191 | 0.51 |
ENSDART00000161021
ENSDART00000124134 |
gfral
|
GDNF family receptor alpha like |
| chr16_-_41004731 | 0.51 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr1_+_44127292 | 0.50 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr14_-_413273 | 0.50 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr3_+_1182315 | 0.49 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr5_+_16117871 | 0.49 |
ENSDART00000090657
|
znrf3
|
zinc and ring finger 3 |
| chr23_+_44741500 | 0.48 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr18_+_9615147 | 0.48 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr8_+_20956078 | 0.48 |
ENSDART00000136397
|
si:dkeyp-82a1.2
|
si:dkeyp-82a1.2 |
| chr14_-_4556896 | 0.47 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr10_+_33393829 | 0.47 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr14_+_23717165 | 0.46 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_-_44064842 | 0.46 |
ENSDART00000151541
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr15_-_9272328 | 0.46 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr6_+_49082796 | 0.45 |
ENSDART00000182446
|
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr1_-_55118745 | 0.45 |
ENSDART00000133915
|
sertad2a
|
SERTA domain containing 2a |
| chr14_-_7207961 | 0.45 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr21_-_16114061 | 0.44 |
ENSDART00000035742
|
cyb561a3b
|
cytochrome b561 family, member A3b |
| chr20_-_46362606 | 0.44 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr7_+_23515966 | 0.44 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr19_-_6988837 | 0.43 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr13_+_12299997 | 0.43 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr7_-_71829649 | 0.43 |
ENSDART00000160449
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr14_-_4145594 | 0.42 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr5_-_42904329 | 0.42 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr22_-_30770751 | 0.42 |
ENSDART00000172115
|
AL831726.2
|
|
| chr11_+_38280454 | 0.42 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr25_+_37293312 | 0.41 |
ENSDART00000086737
ENSDART00000161595 |
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr25_-_13703826 | 0.41 |
ENSDART00000163398
|
pla2g15
|
phospholipase A2, group XV |
| chr5_-_38094130 | 0.41 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr25_-_25384045 | 0.40 |
ENSDART00000150631
|
zgc:123278
|
zgc:123278 |
| chr23_+_9522781 | 0.40 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr6_-_30210378 | 0.40 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr7_-_72261721 | 0.40 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr10_-_5581487 | 0.39 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr20_+_23960525 | 0.39 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr17_-_43556415 | 0.39 |
ENSDART00000190102
ENSDART00000193156 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr17_-_49978986 | 0.38 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr20_-_29864390 | 0.38 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr18_-_48550426 | 0.38 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr25_+_5035343 | 0.37 |
ENSDART00000011751
|
parvb
|
parvin, beta |
| chr4_+_19700308 | 0.37 |
ENSDART00000027919
|
pax4
|
paired box 4 |
| chr16_-_51271962 | 0.37 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr19_+_823945 | 0.37 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr23_+_27740788 | 0.36 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr4_+_3980247 | 0.36 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr8_-_40075983 | 0.36 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr14_+_8940326 | 0.36 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr19_+_770300 | 0.36 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr2_+_8779164 | 0.36 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr24_+_40860320 | 0.35 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr13_+_255067 | 0.35 |
ENSDART00000102505
|
foxg1d
|
forkhead box G1d |
| chr21_+_45841731 | 0.34 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr19_-_9882821 | 0.34 |
ENSDART00000147128
|
cacng7a
|
calcium channel, voltage-dependent, gamma subunit 7a |
| chr24_-_25004553 | 0.34 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr6_-_37468971 | 0.34 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa4a+hoxc3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.5 | 1.4 | GO:0097435 | fibril organization(GO:0097435) |
| 0.4 | 1.7 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 2.1 | GO:0042745 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.3 | 0.9 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.3 | 2.3 | GO:0043092 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.3 | 3.1 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 1.0 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 0.7 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 1.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 1.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 1.5 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 0.6 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.2 | 1.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 1.0 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.1 | 0.4 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.1 | 0.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 2.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.7 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.5 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.6 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.1 | 1.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 12.3 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.6 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0060907 | dendritic cell antigen processing and presentation(GO:0002468) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.3 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 1.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 0.4 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.2 | GO:0032677 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 4.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 2.1 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.2 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.6 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.6 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 1.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.3 | GO:0071422 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 1.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.5 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.3 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.8 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) actin crosslink formation(GO:0051764) |
| 0.0 | 1.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 1.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 1.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 1.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.9 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.0 | GO:0072679 | chemokine production(GO:0032602) negative T cell selection(GO:0043383) thymocyte migration(GO:0072679) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.2 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.0 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.6 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 1.4 | GO:0030891 | VCB complex(GO:0030891) |
| 0.2 | 1.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.6 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.8 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 1.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.8 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.6 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 1.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.7 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 1.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 1.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 1.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 1.7 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 0.8 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 1.8 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.2 | 1.0 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 1.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.5 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.2 | 0.7 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 2.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 11.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.4 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 1.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.9 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.0 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.9 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 1.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.7 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.6 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 1.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 2.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 2.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 3.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 2.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |