Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
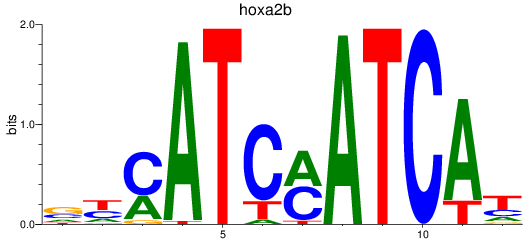
Results for hoxa2b
Z-value: 1.69
Transcription factors associated with hoxa2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa2b
|
ENSDARG00000023031 | homeobox A2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa2b | dr11_v1_chr16_+_20926673_20926673 | -0.91 | 8.4e-08 | Click! |
Activity profile of hoxa2b motif
Sorted Z-values of hoxa2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_1088658 | 10.51 |
ENSDART00000162991
|
BX537249.1
|
|
| chr12_-_11457625 | 10.00 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr18_+_13792490 | 8.57 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr9_-_22135420 | 8.01 |
ENSDART00000184959
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr11_+_36243774 | 7.95 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr9_-_22099536 | 7.33 |
ENSDART00000101923
|
CR391987.1
|
|
| chr7_-_35066457 | 6.85 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr5_+_37837245 | 6.77 |
ENSDART00000171617
|
epd
|
ependymin |
| chr20_-_8443425 | 5.73 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr9_-_22232902 | 5.61 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr22_-_600016 | 5.43 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr9_-_22135576 | 5.34 |
ENSDART00000101902
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr25_+_19105804 | 5.21 |
ENSDART00000104414
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr10_-_20453995 | 5.19 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr14_-_1565317 | 5.17 |
ENSDART00000169496
|
CABZ01064248.1
|
|
| chr15_-_27972474 | 5.12 |
ENSDART00000162753
|
CR391930.1
|
|
| chr24_-_6898302 | 5.04 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr1_+_17676745 | 4.91 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr24_-_40009446 | 4.83 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr12_+_39685485 | 4.68 |
ENSDART00000163403
|
LO017650.1
|
|
| chr7_+_35075847 | 4.52 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr6_-_55585423 | 4.40 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr2_+_33541928 | 4.36 |
ENSDART00000162852
|
BX548164.1
|
|
| chr8_+_22931427 | 4.24 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr25_+_20215964 | 4.13 |
ENSDART00000139235
|
tnnt2d
|
troponin T2d, cardiac |
| chr21_-_43079161 | 4.10 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr9_-_22310919 | 4.05 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr12_-_20362041 | 4.02 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr15_-_12270857 | 3.98 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr9_-_44939104 | 3.93 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr6_+_41186320 | 3.85 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr11_+_25472758 | 3.70 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr6_+_3640381 | 3.70 |
ENSDART00000172078
|
col28a2b
|
collagen, type XXVIII, alpha 2b |
| chr7_+_26326462 | 3.70 |
ENSDART00000173515
|
zanl
|
zonadhesin, like |
| chr15_+_45640906 | 3.62 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr16_-_54455573 | 3.62 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr18_-_898870 | 3.47 |
ENSDART00000151777
ENSDART00000062654 |
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr7_-_4461104 | 3.42 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr17_-_200316 | 3.40 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr24_-_6897884 | 3.39 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr12_-_25916530 | 3.38 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr17_+_40684 | 3.34 |
ENSDART00000164231
|
CABZ01111555.1
|
|
| chr14_+_35237613 | 3.30 |
ENSDART00000163465
|
ebf3a
|
early B cell factor 3a |
| chr8_+_43053519 | 3.29 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr3_+_4997545 | 3.27 |
ENSDART00000181237
|
CABZ01117706.1
|
|
| chr25_+_4760489 | 3.27 |
ENSDART00000167399
|
FP245455.1
|
|
| chr16_+_12022543 | 3.16 |
ENSDART00000012673
|
gnb3a
|
guanine nucleotide binding protein (G protein), beta polypeptide 3a |
| chr25_+_20216159 | 3.14 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr9_+_22003942 | 3.13 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr17_-_6399920 | 3.11 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr20_-_34868814 | 3.09 |
ENSDART00000153049
|
stmn4
|
stathmin-like 4 |
| chr10_+_439692 | 3.04 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr21_+_40092301 | 2.97 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr17_-_20897407 | 2.95 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr8_+_21376290 | 2.94 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr14_-_2199573 | 2.91 |
ENSDART00000124485
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr4_+_21129752 | 2.89 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr16_-_13612650 | 2.88 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr23_+_44881020 | 2.87 |
ENSDART00000149355
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr11_-_7320211 | 2.83 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr18_+_1768804 | 2.82 |
ENSDART00000167477
|
CABZ01088484.1
|
|
| chr4_+_26496489 | 2.76 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr2_-_28671139 | 2.76 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr25_-_35497055 | 2.73 |
ENSDART00000009271
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr21_-_40174647 | 2.72 |
ENSDART00000183738
ENSDART00000076840 ENSDART00000145109 |
slco2b1
|
solute carrier organic anion transporter family, member 2B1 |
| chr13_-_51065048 | 2.71 |
ENSDART00000168085
|
CABZ01078663.1
|
|
| chr25_+_15354095 | 2.54 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr23_-_45504991 | 2.50 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr18_-_42785469 | 2.50 |
ENSDART00000024768
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr5_-_50992690 | 2.49 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr24_-_29822913 | 2.47 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr12_-_4346085 | 2.46 |
ENSDART00000112433
|
ca15c
|
carbonic anhydrase XV c |
| chr22_+_1123873 | 2.44 |
ENSDART00000137708
|
si:ch1073-181h11.2
|
si:ch1073-181h11.2 |
| chr13_+_15838151 | 2.43 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr5_+_44654535 | 2.41 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr1_+_29281764 | 2.38 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr7_+_427503 | 2.37 |
ENSDART00000185942
|
NRXN2 (1 of many)
|
neurexin 2 |
| chr1_-_51465730 | 2.35 |
ENSDART00000074284
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr12_+_48241841 | 2.34 |
ENSDART00000168616
|
ppa1a
|
pyrophosphatase (inorganic) 1a |
| chr2_-_45630823 | 2.34 |
ENSDART00000183553
|
CR450720.1
|
|
| chr4_+_17279966 | 2.27 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr2_+_24199276 | 2.27 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr22_-_33679277 | 2.27 |
ENSDART00000169948
|
FO904977.1
|
|
| chr15_+_45643787 | 2.26 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr19_+_46824723 | 2.25 |
ENSDART00000158620
|
AL953858.1
|
|
| chr2_-_31634978 | 2.24 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr7_-_36096582 | 2.22 |
ENSDART00000188507
|
CR848715.1
|
|
| chr20_-_29420713 | 2.22 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr2_+_17451656 | 2.21 |
ENSDART00000163620
|
BX640408.1
|
|
| chr19_+_4443285 | 2.21 |
ENSDART00000162683
|
trappc9
|
trafficking protein particle complex 9 |
| chr14_+_15155684 | 2.20 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr18_-_7677208 | 2.19 |
ENSDART00000092456
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr12_-_4028079 | 2.16 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr4_-_16333944 | 2.15 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr4_+_40427249 | 2.15 |
ENSDART00000151927
|
si:ch211-218h8.3
|
si:ch211-218h8.3 |
| chr23_+_44611864 | 2.13 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr21_+_11969603 | 2.13 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr4_-_42408339 | 2.11 |
ENSDART00000172612
|
si:ch211-59d8.3
|
si:ch211-59d8.3 |
| chr16_+_10776688 | 2.11 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr22_+_20208185 | 2.11 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr14_-_36378494 | 2.10 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr23_+_5490854 | 2.10 |
ENSDART00000175403
|
tulp1a
|
tubby like protein 1a |
| chr25_+_7494181 | 2.09 |
ENSDART00000165005
|
cat
|
catalase |
| chr10_+_375042 | 2.08 |
ENSDART00000171854
|
si:ch1073-303d10.1
|
si:ch1073-303d10.1 |
| chr17_-_20897250 | 2.08 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr25_-_30047477 | 2.08 |
ENSDART00000164859
|
CR759810.1
|
|
| chr12_+_48634927 | 2.05 |
ENSDART00000168441
|
zgc:165653
|
zgc:165653 |
| chr5_+_36932718 | 2.03 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr15_+_16521785 | 2.02 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr4_+_76575585 | 2.01 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr24_-_29997145 | 2.01 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr10_-_22249444 | 1.99 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr17_-_28749640 | 1.99 |
ENSDART00000000948
|
coch
|
coagulation factor C homolog, cochlin (Limulus polyphemus) |
| chr22_-_24297510 | 1.98 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr15_+_42933236 | 1.97 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr9_+_44721808 | 1.97 |
ENSDART00000190578
|
nckap1
|
NCK-associated protein 1 |
| chr23_+_9867483 | 1.95 |
ENSDART00000023099
|
slc16a7
|
solute carrier family 16, member 7 (monocarboxylic acid transporter 2) |
| chr8_+_49975160 | 1.95 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr5_+_4332220 | 1.95 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr2_-_38000276 | 1.93 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr14_-_31893996 | 1.92 |
ENSDART00000173222
|
gpr101
|
G protein-coupled receptor 101 |
| chr6_+_9175886 | 1.92 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr9_-_51370293 | 1.91 |
ENSDART00000084806
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr2_+_24199073 | 1.90 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr10_-_29809105 | 1.89 |
ENSDART00000162231
|
si:ch73-261i21.5
|
si:ch73-261i21.5 |
| chr2_+_45636034 | 1.89 |
ENSDART00000142230
|
fndc7rs3
|
fibronectin type III domain containing 7, related sequence 3 |
| chr5_-_18446483 | 1.89 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr5_+_32221755 | 1.87 |
ENSDART00000125917
|
myhc4
|
myosin heavy chain 4 |
| chr23_-_16692312 | 1.86 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr25_-_207214 | 1.85 |
ENSDART00000193448
|
FP236318.3
|
|
| chr11_-_4220167 | 1.85 |
ENSDART00000185406
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr13_-_290377 | 1.83 |
ENSDART00000134963
|
chs1
|
chitin synthase 1 |
| chr7_-_13882988 | 1.82 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr17_-_15640467 | 1.81 |
ENSDART00000014210
|
fut9a
|
fucosyltransferase 9a |
| chr25_+_15841670 | 1.80 |
ENSDART00000049992
|
syt9b
|
synaptotagmin IXb |
| chr20_+_23238833 | 1.79 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr25_-_13363286 | 1.79 |
ENSDART00000163735
ENSDART00000169119 |
ndrg4
|
NDRG family member 4 |
| chr2_+_21128391 | 1.76 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr7_-_2129076 | 1.76 |
ENSDART00000182385
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr14_+_2478994 | 1.75 |
ENSDART00000170538
|
CU929317.1
|
|
| chr2_+_52232630 | 1.75 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr14_+_15231097 | 1.75 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr16_+_46111849 | 1.73 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr25_+_19106574 | 1.73 |
ENSDART00000067332
|
rlbp1b
|
retinaldehyde binding protein 1b |
| chr16_-_45058919 | 1.72 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr11_-_44030962 | 1.72 |
ENSDART00000171910
|
FP016005.1
|
|
| chr1_-_43963441 | 1.71 |
ENSDART00000137646
|
odam
|
odontogenic, ameloblast associated |
| chr24_+_5840258 | 1.71 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr18_-_8877077 | 1.71 |
ENSDART00000137266
|
si:dkey-95h12.2
|
si:dkey-95h12.2 |
| chr6_+_4255319 | 1.70 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr22_+_34701848 | 1.70 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr6_+_54538948 | 1.69 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr17_+_5793248 | 1.69 |
ENSDART00000153743
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr8_-_10045043 | 1.67 |
ENSDART00000081917
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr16_-_49746832 | 1.66 |
ENSDART00000166423
|
znf385d
|
zinc finger protein 385D |
| chr1_+_11881559 | 1.65 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr15_-_7598294 | 1.65 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr6_-_16406210 | 1.64 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr15_-_7598542 | 1.63 |
ENSDART00000173092
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr3_-_32818607 | 1.63 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr1_-_26023678 | 1.62 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr13_+_9432501 | 1.61 |
ENSDART00000058064
|
zgc:123321
|
zgc:123321 |
| chr18_+_28102620 | 1.61 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr17_-_26507289 | 1.59 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr4_+_47736069 | 1.58 |
ENSDART00000193329
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr8_-_44015210 | 1.58 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr18_+_783936 | 1.57 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr12_-_26406323 | 1.57 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr19_-_11208782 | 1.54 |
ENSDART00000044426
ENSDART00000189754 |
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr18_+_16963881 | 1.53 |
ENSDART00000147583
|
si:ch211-242e8.1
|
si:ch211-242e8.1 |
| chr8_-_10048059 | 1.52 |
ENSDART00000138411
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr1_-_55044256 | 1.52 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr3_-_48716422 | 1.51 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr19_+_37925616 | 1.51 |
ENSDART00000148348
|
nxph1
|
neurexophilin 1 |
| chr8_-_17771755 | 1.49 |
ENSDART00000063592
|
prkcz
|
protein kinase C, zeta |
| chr18_+_16960966 | 1.48 |
ENSDART00000193969
ENSDART00000100131 |
si:ch211-242e8.1
|
si:ch211-242e8.1 |
| chr8_+_50983551 | 1.48 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr2_-_19109304 | 1.48 |
ENSDART00000168028
|
si:dkey-225f23.5
|
si:dkey-225f23.5 |
| chr13_-_23756700 | 1.48 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr13_-_1349922 | 1.48 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr4_-_14926637 | 1.47 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr24_+_37370064 | 1.46 |
ENSDART00000185870
|
si:ch211-183d21.3
|
si:ch211-183d21.3 |
| chr18_+_10840071 | 1.45 |
ENSDART00000014496
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr8_-_34051548 | 1.45 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr7_+_41295974 | 1.45 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr12_-_3453589 | 1.45 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr12_+_6195191 | 1.44 |
ENSDART00000043236
ENSDART00000186420 |
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr16_+_12281032 | 1.43 |
ENSDART00000138638
|
si:dkey-26c10.5
|
si:dkey-26c10.5 |
| chr8_-_31403938 | 1.43 |
ENSDART00000159168
|
nim1k
|
NIM1 serine/threonine protein kinase |
| chr3_-_35800221 | 1.43 |
ENSDART00000031390
|
caskin1
|
CASK interacting protein 1 |
| chr5_+_61301525 | 1.42 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr1_-_9228007 | 1.42 |
ENSDART00000147277
ENSDART00000135219 |
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr9_-_3653259 | 1.41 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr14_+_34486629 | 1.41 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr17_+_8183393 | 1.41 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr8_+_36509885 | 1.41 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr4_+_7391110 | 1.40 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr22_+_15438513 | 1.39 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr7_+_35229645 | 1.39 |
ENSDART00000144327
|
tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr8_-_53198154 | 1.39 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0015868 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 1.1 | 8.6 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 1.0 | 3.9 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 1.0 | 4.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.7 | 5.7 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.7 | 2.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.6 | 2.4 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.6 | 1.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.6 | 5.8 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.6 | 2.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.5 | 5.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.5 | 1.9 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.4 | 5.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.4 | 0.4 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 0.4 | 2.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.4 | 2.4 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.4 | 1.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.4 | 1.5 | GO:0090330 | regulation of platelet activation(GO:0010543) regulation of platelet aggregation(GO:0090330) |
| 0.4 | 1.5 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.4 | 1.8 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.4 | 9.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.4 | 1.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.4 | 2.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.3 | 1.4 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.3 | 9.1 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.3 | 3.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 1.3 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.3 | 2.2 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.3 | 1.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 1.2 | GO:0006837 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.3 | 0.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.3 | 0.3 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) |
| 0.3 | 8.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.3 | 2.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 0.8 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 3.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 1.0 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.7 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.2 | 2.8 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.2 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 1.4 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.2 | 0.7 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.2 | 0.7 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.2 | 2.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 1.2 | GO:0032648 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.0 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.2 | 1.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 1.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.2 | 3.7 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.2 | 2.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 4.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 1.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 25.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.2 | 0.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 1.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 1.3 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 2.5 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 1.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 8.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 2.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 0.5 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 0.5 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 8.7 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.2 | 0.7 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 2.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 1.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 4.8 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.2 | 0.8 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.2 | 1.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 2.0 | GO:0046349 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.1 | 1.6 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 | 0.9 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 1.6 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.1 | 1.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 3.1 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.7 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 3.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.8 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 1.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 3.3 | GO:0051283 | sequestering of calcium ion(GO:0051208) release of sequestered calcium ion into cytosol(GO:0051209) regulation of sequestering of calcium ion(GO:0051282) negative regulation of sequestering of calcium ion(GO:0051283) |
| 0.1 | 2.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.5 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.4 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 2.7 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 2.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 1.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.4 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 5.3 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 3.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.8 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 1.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.5 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.1 | 3.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 2.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.5 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 0.6 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.8 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 1.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.2 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.7 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 0.4 | GO:0009118 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.1 | 0.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 3.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 2.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.5 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.1 | 2.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 1.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 1.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 8.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 3.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 3.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 1.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 2.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 2.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.2 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.5 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 1.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.7 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.4 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 2.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.6 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0046323 | glucose import(GO:0046323) |
| 0.1 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 1.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 1.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 3.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 5.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 3.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.5 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 1.4 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.8 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.4 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 1.1 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.7 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 2.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 0.3 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 2.0 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.5 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.3 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 1.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0009261 | purine ribonucleotide catabolic process(GO:0009154) ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.5 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 1.3 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 1.1 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 2.1 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.7 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.0 | 0.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0051039 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 1.4 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.4 | GO:0050679 | positive regulation of epithelial cell proliferation(GO:0050679) |
| 0.0 | 0.6 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 2.4 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.2 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 1.7 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.6 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.0 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.6 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0031056 | regulation of histone modification(GO:0031056) |
| 0.0 | 0.0 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.3 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.4 | GO:0071216 | cellular response to biotic stimulus(GO:0071216) cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.5 | GO:1901214 | regulation of neuron death(GO:1901214) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.8 | 3.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.7 | 4.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.6 | 2.9 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.4 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.4 | 2.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 2.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 14.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 1.8 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.3 | 3.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.0 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 1.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 1.5 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 9.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.6 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 1.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 9.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 1.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 4.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 2.2 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 5.5 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 5.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 8.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 9.4 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.6 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 1.3 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 1.7 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 1.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 5.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.2 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.5 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 3.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 22.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 16.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.2 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.4 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 1.1 | 3.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.0 | 4.8 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.8 | 3.3 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.8 | 2.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.7 | 2.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.7 | 2.7 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.7 | 7.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.6 | 2.5 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.6 | 3.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.6 | 1.8 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.6 | 5.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.6 | 2.3 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 4.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 2.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 7.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.4 | 1.7 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.4 | 3.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.4 | 11.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 1.2 | GO:0005335 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 2.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.4 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 1.4 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.3 | 1.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 27.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 1.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.3 | 2.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 0.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 0.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.3 | 2.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.3 | 2.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.3 | 1.1 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.3 | 4.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.3 | 1.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.3 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.2 | 2.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 2.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 2.7 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.2 | 0.7 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.2 | 2.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 3.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.2 | 1.0 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.2 | 0.8 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.2 | 0.6 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.2 | 3.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 0.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 1.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.2 | 3.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 8.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 0.7 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.2 | 1.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.2 | 0.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 6.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 12.9 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.2 | 1.6 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 1.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 2.0 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.2 | 1.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 1.6 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.6 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 4.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.7 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 5.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 2.7 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.5 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.6 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 1.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 8.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 5.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.1 | 0.9 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.8 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.3 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.3 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.5 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.4 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 1.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 17.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 9.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 2.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.3 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.0 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.4 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 1.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.7 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 7.5 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 8.2 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) protein complex scaffold(GO:0032947) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.9 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.5 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 1.9 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 7.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.0 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 3.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.4 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 2.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.3 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 2.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.6 | 3.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.4 | 2.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 8.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.4 | 8.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.3 | 2.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.2 | 2.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 0.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 2.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 0.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 3.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 2.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.9 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 4.9 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 0.6 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 3.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.9 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |