Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
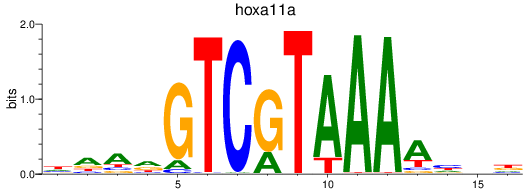
Results for hoxa11a
Z-value: 0.37
Transcription factors associated with hoxa11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa11a
|
ENSDARG00000104162 | homeobox A11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa11a | dr11_v1_chr19_+_19737214_19737300 | 0.69 | 1.2e-03 | Click! |
Activity profile of hoxa11a motif
Sorted Z-values of hoxa11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_13128180 | 0.92 |
ENSDART00000081426
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr13_-_25196758 | 0.69 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr11_+_2198831 | 0.59 |
ENSDART00000160515
|
hoxc6b
|
homeobox C6b |
| chr12_-_26851726 | 0.47 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr24_-_21090447 | 0.46 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr23_+_36087219 | 0.45 |
ENSDART00000154825
|
hoxc3a
|
homeobox C3a |
| chr21_+_13127742 | 0.43 |
ENSDART00000179221
|
odf2a
|
outer dense fiber of sperm tails 2a |
| chr7_+_20524064 | 0.42 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr3_-_40254634 | 0.41 |
ENSDART00000154562
|
top3a
|
DNA topoisomerase III alpha |
| chr1_-_462165 | 0.39 |
ENSDART00000152799
|
si:ch73-244f7.3
|
si:ch73-244f7.3 |
| chr19_+_7424347 | 0.36 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr5_-_31857345 | 0.34 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr2_-_6065416 | 0.33 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr2_-_43851915 | 0.32 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr7_-_18508815 | 0.32 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr9_-_3671911 | 0.31 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr15_+_8767650 | 0.31 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr19_-_22507715 | 0.29 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr1_+_45056371 | 0.27 |
ENSDART00000073689
ENSDART00000167309 |
btr01
|
bloodthirsty-related gene family, member 1 |
| chr20_-_37820939 | 0.27 |
ENSDART00000032978
|
nsl1
|
NSL1, MIS12 kinetochore complex component |
| chr7_+_20966434 | 0.27 |
ENSDART00000185570
|
efnb3b
|
ephrin-B3b |
| chr18_-_2433011 | 0.27 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr15_+_9297340 | 0.25 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr22_+_2239974 | 0.25 |
ENSDART00000141993
|
znf1144
|
zinc finger protein 1144 |
| chr5_-_31856681 | 0.24 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr12_+_27117609 | 0.24 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr12_-_11560794 | 0.24 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr20_+_43379029 | 0.24 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr3_+_36284986 | 0.24 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr15_+_44093286 | 0.23 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr16_-_6205790 | 0.22 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr4_+_39368978 | 0.22 |
ENSDART00000160640
|
si:dkey-261o4.1
|
si:dkey-261o4.1 |
| chr2_-_43852207 | 0.22 |
ENSDART00000192627
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr9_-_2892045 | 0.22 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr12_+_33361948 | 0.20 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr24_+_21540842 | 0.20 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr1_+_36722122 | 0.20 |
ENSDART00000111566
|
tmem184c
|
transmembrane protein 184C |
| chr13_+_35472803 | 0.19 |
ENSDART00000011583
|
mkks
|
McKusick-Kaufman syndrome |
| chr11_+_24716837 | 0.19 |
ENSDART00000145217
|
zgc:153953
|
zgc:153953 |
| chr16_+_20910186 | 0.19 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr4_-_20108833 | 0.19 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr12_-_34713690 | 0.19 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr14_-_48348973 | 0.19 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr20_+_34671386 | 0.18 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr5_+_59397348 | 0.18 |
ENSDART00000175642
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr8_-_12432604 | 0.18 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr5_+_59397739 | 0.18 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr13_+_42309688 | 0.17 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr6_+_49551614 | 0.17 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr2_+_43851983 | 0.17 |
ENSDART00000126413
|
nlrb5
|
NOD-like receptor family B, member 5 |
| chr18_-_17087138 | 0.16 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr6_+_33885828 | 0.16 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr9_-_2892250 | 0.16 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr25_-_18002498 | 0.16 |
ENSDART00000158688
|
cep290
|
centrosomal protein 290 |
| chr11_+_25560632 | 0.15 |
ENSDART00000033914
|
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr12_-_34713888 | 0.15 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr17_+_17764979 | 0.15 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr2_-_13254821 | 0.13 |
ENSDART00000022621
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr23_-_1658980 | 0.13 |
ENSDART00000186227
|
CU693481.1
|
|
| chr17_-_31695217 | 0.12 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr20_+_37820992 | 0.12 |
ENSDART00000064692
|
tatdn3
|
TatD DNase domain containing 3 |
| chr7_-_30624435 | 0.11 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr3_-_18737126 | 0.11 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr24_-_31223232 | 0.11 |
ENSDART00000164155
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr13_+_45431660 | 0.10 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr17_+_4368859 | 0.09 |
ENSDART00000055385
|
crls1
|
cardiolipin synthase 1 |
| chr6_+_3730843 | 0.09 |
ENSDART00000019630
|
FO704755.1
|
|
| chr14_-_31060082 | 0.09 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr13_-_9875538 | 0.08 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr24_+_20658942 | 0.07 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr13_+_29925397 | 0.07 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr25_-_18002937 | 0.07 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr20_-_46541834 | 0.07 |
ENSDART00000060685
ENSDART00000181720 |
tmed10
|
transmembrane p24 trafficking protein 10 |
| chr2_-_3158919 | 0.06 |
ENSDART00000098394
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr11_+_25560072 | 0.06 |
ENSDART00000124131
ENSDART00000147179 |
mbd1b
|
methyl-CpG binding domain protein 1b |
| chr24_-_40009446 | 0.06 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr16_-_47426482 | 0.06 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr24_+_20658760 | 0.05 |
ENSDART00000188362
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr9_-_29039506 | 0.05 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr11_+_41135055 | 0.05 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr1_-_29061285 | 0.04 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr8_+_23861461 | 0.04 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr2_-_13254594 | 0.04 |
ENSDART00000155671
|
kdsr
|
3-ketodihydrosphingosine reductase |
| chr2_-_9259283 | 0.04 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr17_+_44463230 | 0.04 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr11_-_40257225 | 0.04 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr22_+_10646928 | 0.03 |
ENSDART00000038465
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr19_-_33370271 | 0.03 |
ENSDART00000132628
|
nkd3l
|
naked cuticle homolog 3, like |
| chr7_+_9308625 | 0.03 |
ENSDART00000084598
|
selenos
|
selenoprotein S |
| chr16_-_10316359 | 0.03 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr6_-_53334259 | 0.03 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr15_-_14552101 | 0.01 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr5_+_32932357 | 0.00 |
ENSDART00000192397
|
lmo4a
|
LIM domain only 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.2 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.3 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |