Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
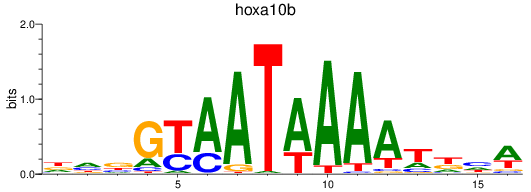
Results for hoxa10b
Z-value: 0.94
Transcription factors associated with hoxa10b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa10b
|
ENSDARG00000031337 | homeobox A10b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa10b | dr11_v1_chr16_+_20910186_20910186 | 0.88 | 6.6e-07 | Click! |
Activity profile of hoxa10b motif
Sorted Z-values of hoxa10b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_29499363 | 2.81 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_-_47571456 | 2.48 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr11_+_25257022 | 2.33 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr19_-_47571797 | 2.31 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr11_-_25257045 | 2.30 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr9_+_30090656 | 2.27 |
ENSDART00000102981
|
col8a1a
|
collagen, type VIII, alpha 1a |
| chr16_-_27566552 | 1.76 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr14_-_32965169 | 1.73 |
ENSDART00000114973
|
cdx4
|
caudal type homeobox 4 |
| chr1_-_53468160 | 1.70 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr2_-_10877765 | 1.70 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr3_+_6443992 | 1.59 |
ENSDART00000169325
ENSDART00000162255 |
nup85
|
nucleoporin 85 |
| chr14_-_16082806 | 1.58 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr8_+_39795918 | 1.50 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr5_+_70155935 | 1.46 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr17_+_43595692 | 1.45 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr18_+_9171778 | 1.41 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr3_-_58650057 | 1.41 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr2_-_50272077 | 1.40 |
ENSDART00000127623
|
cul1a
|
cullin 1a |
| chr22_+_10606573 | 1.39 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr1_-_14506759 | 1.38 |
ENSDART00000057044
|
si:dkey-194g4.1
|
si:dkey-194g4.1 |
| chr5_-_31856681 | 1.37 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr21_-_11970199 | 1.37 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr3_+_31093455 | 1.36 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr24_-_21090447 | 1.36 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr20_-_2641233 | 1.36 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr2_-_17115256 | 1.35 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr14_+_10625112 | 1.31 |
ENSDART00000143377
ENSDART00000136480 |
nup62l
|
nucleoporin 62 like |
| chr22_+_15959844 | 1.30 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr16_+_52966812 | 1.26 |
ENSDART00000148435
ENSDART00000049099 ENSDART00000150117 |
trip13
|
thyroid hormone receptor interactor 13 |
| chr10_-_21545091 | 1.26 |
ENSDART00000029122
ENSDART00000132207 |
zgc:165539
|
zgc:165539 |
| chr14_+_26439227 | 1.22 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr10_-_34889053 | 1.21 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr4_+_13428993 | 1.21 |
ENSDART00000067151
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| chr5_-_31857345 | 1.19 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr19_+_7424347 | 1.13 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr25_-_29074064 | 1.12 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr24_-_15159658 | 1.11 |
ENSDART00000142473
|
rttn
|
rotatin |
| chr9_+_3429662 | 1.10 |
ENSDART00000160977
ENSDART00000114168 ENSDART00000082153 |
CU469503.1
itga6a
|
integrin, alpha 6a |
| chr12_+_19199735 | 1.10 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr7_-_8712148 | 1.10 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr3_-_12026741 | 1.10 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr2_+_20967673 | 1.10 |
ENSDART00000057174
|
arpc5a
|
actin related protein 2/3 complex, subunit 5A |
| chr15_-_5901514 | 1.09 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr25_-_14433503 | 1.08 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr21_-_26071142 | 1.06 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr17_+_15535501 | 1.05 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr20_-_36416922 | 1.05 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr12_-_18961491 | 1.04 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr23_-_21215311 | 1.03 |
ENSDART00000112424
|
megf6a
|
multiple EGF-like-domains 6a |
| chr15_+_30323491 | 1.01 |
ENSDART00000048847
|
nos2b
|
nitric oxide synthase 2b, inducible |
| chr16_+_31942527 | 1.01 |
ENSDART00000188334
ENSDART00000188643 ENSDART00000058496 |
phc1
|
polyhomeotic homolog 1 |
| chr2_-_42035250 | 1.01 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr21_+_39292266 | 0.99 |
ENSDART00000144116
|
si:ch211-274p24.2
|
si:ch211-274p24.2 |
| chr6_+_40554551 | 0.99 |
ENSDART00000017859
ENSDART00000155928 |
ddi2
|
DNA-damage inducible protein 2 |
| chr4_-_22749553 | 0.98 |
ENSDART00000040033
|
nup107
|
nucleoporin 107 |
| chr8_+_23105117 | 0.96 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr24_-_3407507 | 0.95 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr23_-_29751730 | 0.93 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr21_+_19334198 | 0.93 |
ENSDART00000147372
|
helq
|
helicase, POLQ like |
| chr12_-_10476448 | 0.93 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr11_-_25257595 | 0.93 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr8_-_31416403 | 0.93 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr12_+_3262564 | 0.92 |
ENSDART00000184264
|
tmem101
|
transmembrane protein 101 |
| chr4_-_61651223 | 0.92 |
ENSDART00000172688
|
si:dkey-26i24.1
|
si:dkey-26i24.1 |
| chr15_+_23534126 | 0.91 |
ENSDART00000152320
|
si:dkey-182i3.10
|
si:dkey-182i3.10 |
| chr3_+_60589157 | 0.91 |
ENSDART00000165367
|
mettl23
|
methyltransferase like 23 |
| chr25_+_3549841 | 0.91 |
ENSDART00000164030
|
ccdc77
|
coiled-coil domain containing 77 |
| chr6_-_7107868 | 0.91 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr7_+_51103416 | 0.89 |
ENSDART00000174236
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr12_+_19191787 | 0.89 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr12_+_16452575 | 0.89 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr1_-_12394048 | 0.89 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr2_-_20715094 | 0.88 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr13_+_1131748 | 0.88 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr6_-_10728921 | 0.88 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr6_+_54221654 | 0.88 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr17_+_17764979 | 0.87 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr7_-_16582187 | 0.87 |
ENSDART00000131726
|
e2f8
|
E2F transcription factor 8 |
| chr12_-_5728755 | 0.87 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr7_+_34290051 | 0.86 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr11_-_18323059 | 0.86 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr1_-_8566567 | 0.85 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr22_+_2512154 | 0.84 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr12_+_16452912 | 0.83 |
ENSDART00000192467
|
kif20bb
|
kinesin family member 20Bb |
| chr20_+_26943072 | 0.83 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr10_+_18877362 | 0.82 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr21_-_26071773 | 0.81 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr22_+_1947494 | 0.81 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr22_-_14115292 | 0.81 |
ENSDART00000105717
ENSDART00000165670 |
aox5
|
aldehyde oxidase 5 |
| chr23_-_21215520 | 0.81 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr2_+_19163965 | 0.81 |
ENSDART00000166073
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr13_-_24717618 | 0.80 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr20_+_6659770 | 0.79 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr24_-_4450238 | 0.78 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr11_-_25461336 | 0.78 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr25_-_3759322 | 0.78 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr2_+_21048661 | 0.77 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr7_+_51103232 | 0.77 |
ENSDART00000073827
|
col4a5
|
collagen, type IV, alpha 5 (Alport syndrome) |
| chr4_-_39171837 | 0.75 |
ENSDART00000179734
|
si:ch211-22k7.9
|
si:ch211-22k7.9 |
| chr4_-_17353972 | 0.75 |
ENSDART00000041529
|
parpbp
|
PARP1 binding protein |
| chr12_+_13205955 | 0.75 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr25_+_9013342 | 0.74 |
ENSDART00000154207
ENSDART00000153705 |
im:7145024
|
im:7145024 |
| chr21_+_25625026 | 0.74 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr12_+_26670778 | 0.74 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr4_+_17353714 | 0.74 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr3_-_45778123 | 0.74 |
ENSDART00000146211
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr18_+_6857071 | 0.73 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr12_-_18961289 | 0.73 |
ENSDART00000168405
|
ep300a
|
E1A binding protein p300 a |
| chr8_-_11324143 | 0.73 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr12_-_16452200 | 0.73 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr6_+_11989537 | 0.73 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr1_+_41596099 | 0.73 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr22_+_1953096 | 0.72 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr14_+_34966598 | 0.72 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr21_+_43702016 | 0.72 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr21_+_43404945 | 0.71 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr24_+_19578935 | 0.70 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr21_-_3007412 | 0.70 |
ENSDART00000190839
|
CKS2
|
zgc:86839 |
| chr7_+_15308219 | 0.70 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr12_-_33817114 | 0.69 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr25_+_3549401 | 0.69 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr21_+_28478663 | 0.68 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr25_-_31171948 | 0.68 |
ENSDART00000184275
|
FQ976913.1
|
|
| chr20_+_17581881 | 0.68 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr12_+_3871452 | 0.68 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr12_+_33361948 | 0.68 |
ENSDART00000124982
|
fasn
|
fatty acid synthase |
| chr11_-_5953636 | 0.67 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr1_-_13989643 | 0.67 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr17_-_43666166 | 0.67 |
ENSDART00000077990
|
egr2a
|
early growth response 2a |
| chr13_+_9559461 | 0.66 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr4_+_23125689 | 0.66 |
ENSDART00000077854
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr13_-_7575216 | 0.65 |
ENSDART00000159443
|
pitx3
|
paired-like homeodomain 3 |
| chr12_-_20409794 | 0.64 |
ENSDART00000077936
|
lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr5_-_65121747 | 0.64 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr6_+_35362225 | 0.64 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr3_+_39579393 | 0.64 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr13_+_24717880 | 0.64 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr3_-_5277041 | 0.64 |
ENSDART00000046995
|
txn2
|
thioredoxin 2 |
| chr12_-_4243268 | 0.64 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr12_+_11352630 | 0.63 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr15_-_19772372 | 0.62 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr11_+_6010177 | 0.62 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr24_+_14581864 | 0.61 |
ENSDART00000134536
|
thtpa
|
thiamine triphosphatase |
| chr3_+_13600714 | 0.61 |
ENSDART00000162124
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr8_-_16788626 | 0.60 |
ENSDART00000191652
|
CR759968.2
|
|
| chr2_-_37458527 | 0.60 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr19_+_636886 | 0.59 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr20_-_34670236 | 0.59 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr20_+_25586099 | 0.58 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr15_-_1022436 | 0.58 |
ENSDART00000156003
|
znf1010
|
zinc finger protein 1010 |
| chr9_-_31278048 | 0.58 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr1_-_40911332 | 0.58 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr13_-_24717365 | 0.58 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr15_-_21132480 | 0.57 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr8_-_38159805 | 0.57 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr7_+_22691961 | 0.57 |
ENSDART00000041615
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr22_+_2403068 | 0.56 |
ENSDART00000132925
ENSDART00000132569 |
zgc:112977
|
zgc:112977 |
| chr17_-_36529449 | 0.56 |
ENSDART00000187252
|
colec11
|
collectin sub-family member 11 |
| chr15_+_14202114 | 0.56 |
ENSDART00000169369
|
si:dkey-243k1.3
|
si:dkey-243k1.3 |
| chr2_+_35733335 | 0.55 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr4_-_2265271 | 0.55 |
ENSDART00000125815
|
krr1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr14_-_7409364 | 0.55 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr1_-_9485939 | 0.55 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr13_+_8892784 | 0.54 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr24_+_14541013 | 0.54 |
ENSDART00000066721
|
ngdn
|
neuroguidin, EIF4E binding protein |
| chr17_-_25630822 | 0.54 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr3_+_24275597 | 0.54 |
ENSDART00000183926
ENSDART00000169765 |
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr14_+_22132388 | 0.53 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr18_+_20532618 | 0.53 |
ENSDART00000006109
|
agk
|
acylglycerol kinase |
| chr22_-_15957041 | 0.53 |
ENSDART00000149236
ENSDART00000187500 ENSDART00000176304 ENSDART00000080047 ENSDART00000190068 |
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr2_+_16798923 | 0.53 |
ENSDART00000181603
ENSDART00000179816 ENSDART00000087107 ENSDART00000187009 |
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr24_+_23791758 | 0.52 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr11_+_11303458 | 0.52 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr11_-_23314242 | 0.52 |
ENSDART00000182394
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr3_-_4501026 | 0.52 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr3_+_40576447 | 0.52 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr3_-_60589292 | 0.52 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr20_-_34768301 | 0.51 |
ENSDART00000142482
|
paqr8
|
progestin and adipoQ receptor family member VIII |
| chr1_-_54735441 | 0.51 |
ENSDART00000109089
|
cdc25d
|
cell division cycle 25 homolog d |
| chr1_+_45663727 | 0.51 |
ENSDART00000038574
ENSDART00000141144 ENSDART00000149565 |
trappc5
|
trafficking protein particle complex 5 |
| chr23_+_43718115 | 0.51 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr2_+_25315591 | 0.50 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr1_+_27808916 | 0.50 |
ENSDART00000102335
|
pspc1
|
paraspeckle component 1 |
| chr24_-_20926812 | 0.49 |
ENSDART00000130958
|
ccdc58
|
coiled-coil domain containing 58 |
| chr12_+_23912074 | 0.49 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr1_-_41374117 | 0.48 |
ENSDART00000074777
|
htt
|
huntingtin |
| chr23_-_39959173 | 0.48 |
ENSDART00000077122
|
xcr1a.1
|
chemokine (C motif) receptor 1a, duplicate 1 |
| chr21_-_2299002 | 0.48 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr23_+_20431140 | 0.48 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr19_-_26964348 | 0.47 |
ENSDART00000103955
|
zbtb12.2
|
zinc finger and BTB domain containing 12, tandem duplicate 2 |
| chr5_+_20453874 | 0.47 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr1_+_57371447 | 0.47 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr1_-_38908197 | 0.47 |
ENSDART00000101489
|
vegfc
|
vascular endothelial growth factor c |
| chr25_+_28823952 | 0.47 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr2_-_49627099 | 0.47 |
ENSDART00000083710
ENSDART00000147421 |
cactin
|
cactin |
| chr23_+_12455295 | 0.46 |
ENSDART00000143752
ENSDART00000135785 |
si:ch211-153a8.4
|
si:ch211-153a8.4 |
| chr5_-_66215928 | 0.46 |
ENSDART00000082797
|
coq2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr1_+_46194333 | 0.46 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr15_-_47857687 | 0.46 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr14_+_18785727 | 0.46 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa10b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 1.7 | GO:0007440 | foregut morphogenesis(GO:0007440) |
| 0.3 | 1.7 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 1.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.3 | 1.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.3 | 0.9 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.5 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.3 | 0.9 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.4 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.3 | 0.8 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.2 | 0.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.7 | GO:0021577 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.2 | 0.9 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.2 | 2.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 0.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 1.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 0.6 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.2 | 0.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.2 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.5 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 0.9 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.2 | 2.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.9 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 1.0 | GO:0030826 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.2 | 3.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 1.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.7 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.5 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.7 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.4 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.6 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 1.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.4 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.5 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.5 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.6 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 1.0 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.1 | 0.5 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 2.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.8 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.0 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 0.6 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 1.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.5 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.1 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.6 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.9 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 1.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.2 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.6 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.8 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.7 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.9 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 2.6 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 1.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.5 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.7 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.9 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.6 | GO:0071241 | cellular response to inorganic substance(GO:0071241) |
| 0.0 | 0.5 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0033555 | neuronal action potential(GO:0019228) multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.1 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.2 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 2.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.5 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.7 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.4 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.9 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 0.0 | GO:0051012 | microtubule sliding(GO:0051012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.6 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 0.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 0.9 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 0.7 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.2 | 0.5 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 1.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.6 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.5 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 2.6 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.4 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 2.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 4.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 2.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.4 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.0 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.8 | 3.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.4 | 0.8 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.3 | 1.3 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.9 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.2 | 0.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 0.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.5 | GO:0001729 | ceramide kinase activity(GO:0001729) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.5 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.2 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 0.5 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.2 | 0.5 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.2 | 0.6 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 1.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 3.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 2.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 1.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.2 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 2.7 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 1.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 2.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.8 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 3.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.7 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.0 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.0 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 3.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.3 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.7 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |