Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
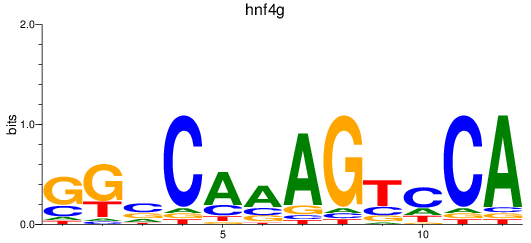
Results for hnf4g
Z-value: 0.65
Transcription factors associated with hnf4g
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf4g
|
ENSDARG00000071565 | hepatocyte nuclear factor 4, gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf4g | dr11_v1_chr24_-_23671709_23671709 | 0.58 | 9.4e-03 | Click! |
Activity profile of hnf4g motif
Sorted Z-values of hnf4g motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_1138802 | 2.01 |
ENSDART00000191130
ENSDART00000187308 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr9_+_1138323 | 1.79 |
ENSDART00000190352
ENSDART00000190387 |
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr20_-_25522911 | 1.72 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr8_-_20838342 | 1.55 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr22_-_31517300 | 1.43 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr17_+_50701748 | 1.43 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr13_-_303137 | 1.37 |
ENSDART00000099131
|
chs1
|
chitin synthase 1 |
| chr9_-_44953664 | 1.28 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr23_+_19790962 | 1.27 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr12_+_22607761 | 1.05 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr2_+_37897079 | 1.01 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr23_-_43595956 | 0.98 |
ENSDART00000162186
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr10_+_45128375 | 0.98 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr6_+_59029485 | 0.85 |
ENSDART00000050140
|
CABZ01088367.1
|
|
| chr12_-_33706726 | 0.83 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr8_+_53464216 | 0.80 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr15_-_41700406 | 0.77 |
ENSDART00000181460
|
si:ch211-276c2.4
|
si:ch211-276c2.4 |
| chr24_-_32150276 | 0.75 |
ENSDART00000166212
|
cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr3_+_16762483 | 0.72 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr2_+_11670270 | 0.70 |
ENSDART00000100524
|
ftr01
|
finTRIM family, member 1 |
| chr8_+_14778292 | 0.70 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr6_-_607063 | 0.68 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr17_+_20173882 | 0.67 |
ENSDART00000155379
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr21_-_43474012 | 0.66 |
ENSDART00000065104
|
tmem185
|
transmembrane protein 185 |
| chr3_-_27868183 | 0.64 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr7_-_24520866 | 0.61 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr18_-_9016161 | 0.59 |
ENSDART00000189888
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr20_+_43379029 | 0.59 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr4_+_7888047 | 0.58 |
ENSDART00000104676
|
camk1da
|
calcium/calmodulin-dependent protein kinase 1Da |
| chr5_-_69180587 | 0.57 |
ENSDART00000156681
ENSDART00000160753 |
zgc:171967
|
zgc:171967 |
| chr13_-_50309969 | 0.56 |
ENSDART00000191597
|
CU928131.1
|
|
| chr19_-_22387141 | 0.54 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr1_+_47585700 | 0.54 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr17_-_6076266 | 0.53 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr17_-_6076084 | 0.51 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr6_-_46474483 | 0.46 |
ENSDART00000155761
|
rdh20
|
retinol dehydrogenase 20 |
| chr5_+_42467867 | 0.46 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr22_+_2844865 | 0.43 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr8_+_47188154 | 0.43 |
ENSDART00000137319
|
si:dkeyp-100a1.6
|
si:dkeyp-100a1.6 |
| chr17_+_20174044 | 0.41 |
ENSDART00000156028
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr13_+_50375800 | 0.41 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr13_-_37254777 | 0.41 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr21_-_27443995 | 0.40 |
ENSDART00000003508
|
bfb
|
complement component bfb |
| chr14_-_16810401 | 0.40 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr4_-_17725008 | 0.40 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr19_+_1003332 | 0.39 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr20_-_46694573 | 0.39 |
ENSDART00000182557
|
AL929435.2
|
|
| chr6_+_27624023 | 0.39 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr16_+_11623956 | 0.39 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr9_+_48415043 | 0.39 |
ENSDART00000159930
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr20_-_49657134 | 0.38 |
ENSDART00000151248
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr11_+_33284837 | 0.38 |
ENSDART00000166239
ENSDART00000111412 |
nkpd1
|
NTPase, KAP family P-loop domain containing 1 |
| chr1_-_37383741 | 0.38 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr16_+_11660839 | 0.38 |
ENSDART00000193911
ENSDART00000143683 |
si:dkey-250k15.10
|
si:dkey-250k15.10 |
| chr11_-_5563498 | 0.37 |
ENSDART00000160835
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr16_+_9495583 | 0.36 |
ENSDART00000150750
ENSDART00000150457 |
pimr208
|
Pim proto-oncogene, serine/threonine kinase, related 208 |
| chr5_-_69180227 | 0.35 |
ENSDART00000154816
|
zgc:171967
|
zgc:171967 |
| chr7_-_29115772 | 0.34 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr21_-_5879897 | 0.34 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr7_+_30823749 | 0.33 |
ENSDART00000085661
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr15_-_30857350 | 0.33 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr21_-_11856143 | 0.33 |
ENSDART00000151204
|
ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr24_-_39567 | 0.33 |
ENSDART00000055488
|
cdh7b
|
cadherin 7b |
| chr19_-_28360033 | 0.32 |
ENSDART00000186994
|
si:dkey-261i16.5
|
si:dkey-261i16.5 |
| chr6_-_49769100 | 0.32 |
ENSDART00000148853
|
si:dkey-249d8.1
|
si:dkey-249d8.1 |
| chr22_-_20376488 | 0.31 |
ENSDART00000140187
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr8_-_49207319 | 0.30 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr14_+_9009600 | 0.28 |
ENSDART00000133904
|
si:ch211-274f20.2
|
si:ch211-274f20.2 |
| chr5_+_37069605 | 0.28 |
ENSDART00000161977
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr3_-_8765165 | 0.27 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr9_+_29994439 | 0.27 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr13_-_40726865 | 0.27 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr10_-_43568239 | 0.26 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr1_-_37383539 | 0.26 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr11_+_11152214 | 0.25 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr5_-_30080332 | 0.25 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr6_-_56103718 | 0.24 |
ENSDART00000191002
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr15_-_2632891 | 0.24 |
ENSDART00000081840
|
cldnj
|
claudin j |
| chr7_+_24520518 | 0.23 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr10_-_21953643 | 0.23 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
| chr18_+_33725576 | 0.22 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr6_+_27339962 | 0.22 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr13_+_21779975 | 0.22 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr22_+_1300587 | 0.21 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr7_-_59514547 | 0.21 |
ENSDART00000168457
|
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr16_+_5251768 | 0.20 |
ENSDART00000144558
|
plecb
|
plectin b |
| chr5_-_65319387 | 0.19 |
ENSDART00000164649
|
col27a1b
|
collagen, type XXVII, alpha 1b |
| chr10_+_41765944 | 0.19 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
| chr21_-_43428040 | 0.19 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr9_+_29616854 | 0.19 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr12_+_2455429 | 0.18 |
ENSDART00000182929
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr20_+_23238833 | 0.17 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr2_+_13050225 | 0.15 |
ENSDART00000182237
|
BX324233.1
|
|
| chr15_-_16012963 | 0.14 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr10_-_39055620 | 0.13 |
ENSDART00000187437
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr4_-_170120 | 0.13 |
ENSDART00000171333
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr14_-_16379843 | 0.13 |
ENSDART00000090711
|
ltc4s
|
leukotriene C4 synthase |
| chr5_-_62317496 | 0.13 |
ENSDART00000180089
|
zgc:85789
|
zgc:85789 |
| chr1_-_52292235 | 0.13 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr22_+_19407531 | 0.12 |
ENSDART00000141060
|
si:dkey-78l4.2
|
si:dkey-78l4.2 |
| chr7_+_17229282 | 0.12 |
ENSDART00000097982
|
slc6a5
|
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr3_+_12764894 | 0.11 |
ENSDART00000166071
|
cyp2k16
|
cytochrome P450, family 2, subfamily K, polypeptide16 |
| chr17_+_15297398 | 0.11 |
ENSDART00000156574
|
si:ch211-270g19.5
|
si:ch211-270g19.5 |
| chr11_+_19068442 | 0.10 |
ENSDART00000171766
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr6_+_11026866 | 0.10 |
ENSDART00000189619
|
abcc6a
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6a |
| chr1_+_41690402 | 0.09 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr4_-_72468168 | 0.09 |
ENSDART00000182995
ENSDART00000174067 |
CR788316.1
|
|
| chr5_-_50640171 | 0.09 |
ENSDART00000183353
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr4_+_11172964 | 0.09 |
ENSDART00000193598
ENSDART00000138661 |
tspan11
|
tetraspanin 11 |
| chr1_-_57172294 | 0.07 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr24_+_20960216 | 0.07 |
ENSDART00000133008
|
si:ch211-161h7.8
|
si:ch211-161h7.8 |
| chr15_+_3825117 | 0.07 |
ENSDART00000183315
|
CABZ01061591.1
|
|
| chr8_+_23390844 | 0.05 |
ENSDART00000184556
|
si:dkey-16n15.6
|
si:dkey-16n15.6 |
| chr16_+_25809235 | 0.04 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr7_-_39738460 | 0.03 |
ENSDART00000052201
|
ccdc96
|
coiled-coil domain containing 96 |
| chr12_+_33916939 | 0.03 |
ENSDART00000149292
|
trim8b
|
tripartite motif containing 8b |
| chr6_+_18298444 | 0.02 |
ENSDART00000166018
|
card14
|
caspase recruitment domain family, member 14 |
| chr8_+_24745041 | 0.00 |
ENSDART00000148872
|
slc16a4
|
solute carrier family 16, member 4 |
| chr24_+_7828097 | 0.00 |
ENSDART00000134975
|
zgc:101569
|
zgc:101569 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf4g
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.3 | 1.3 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.3 | 1.4 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.6 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.3 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.4 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 1.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.4 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 1.0 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 1.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 1.5 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 1.8 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 1.0 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.2 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.0 | 1.5 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 1.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.0 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.8 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.3 | 1.0 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.2 | 1.4 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.7 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.6 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 1.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |