Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
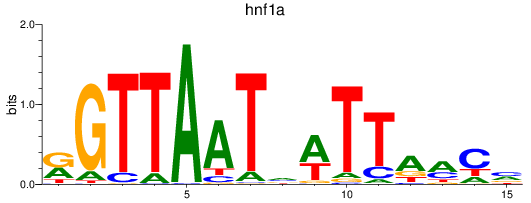
Results for hnf1a
Z-value: 2.51
Transcription factors associated with hnf1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hnf1a
|
ENSDARG00000009470 | HNF1 homeobox a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hnf1a | dr11_v1_chr8_+_39802506_39802506 | 0.07 | 7.9e-01 | Click! |
Activity profile of hnf1a motif
Sorted Z-values of hnf1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_35066457 | 14.42 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr4_+_18804317 | 11.66 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr5_-_30620625 | 10.38 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr24_-_4765740 | 10.27 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr5_+_45677781 | 9.71 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr1_-_59348118 | 9.50 |
ENSDART00000170901
|
cyp3a65
|
cytochrome P450, family 3, subfamily A, polypeptide 65 |
| chr9_-_9982696 | 8.77 |
ENSDART00000192548
ENSDART00000125852 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr9_-_9977827 | 8.17 |
ENSDART00000187315
ENSDART00000010246 |
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr6_-_39344259 | 7.23 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr20_+_35473288 | 7.12 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr6_+_30456788 | 6.10 |
ENSDART00000121492
|
FP236735.1
|
|
| chr7_-_29534001 | 6.03 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr21_+_40092301 | 5.98 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr20_-_40750953 | 5.70 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr3_+_31039923 | 5.28 |
ENSDART00000147706
|
cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr8_-_40555340 | 5.15 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr5_+_28848870 | 5.13 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr12_+_6117682 | 5.02 |
ENSDART00000146650
|
asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr6_+_34028532 | 4.96 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
| chr14_+_31721171 | 4.43 |
ENSDART00000173034
|
adgrg4a
|
adhesion G protein-coupled receptor G4a |
| chr11_-_23332592 | 4.39 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr5_+_9382301 | 4.34 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr5_+_28849155 | 4.24 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr9_+_27339505 | 4.21 |
ENSDART00000011822
|
slc10a2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr7_+_2849020 | 4.16 |
ENSDART00000168695
|
BX004857.1
|
|
| chr5_+_28830643 | 4.15 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr12_-_6172154 | 4.06 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr12_-_6177894 | 3.97 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr4_+_7827261 | 3.79 |
ENSDART00000129568
|
phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr5_+_28830388 | 3.75 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr6_+_38381957 | 3.60 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr2_-_41942666 | 3.54 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr4_-_9533345 | 3.51 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr5_+_9405283 | 3.44 |
ENSDART00000127306
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr13_+_23988442 | 3.43 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr1_-_12126535 | 3.30 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr5_+_9377005 | 3.24 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr16_+_34531486 | 3.08 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr2_-_23778180 | 3.04 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr8_+_16990120 | 3.02 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_37886063 | 2.92 |
ENSDART00000131378
ENSDART00000132152 |
si:ch211-139a5.9
|
si:ch211-139a5.9 |
| chr24_-_39610585 | 2.87 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr6_+_41039166 | 2.87 |
ENSDART00000125659
|
entpd8
|
ectonucleoside triphosphate diphosphohydrolase 8 |
| chr17_-_7371564 | 2.80 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr11_-_37997419 | 2.63 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr12_+_35067863 | 2.55 |
ENSDART00000152849
|
syt15
|
synaptotagmin XV |
| chr14_-_41556720 | 2.49 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr18_+_15644559 | 2.45 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr15_-_23467750 | 2.45 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr5_+_9417409 | 2.45 |
ENSDART00000125421
ENSDART00000130265 |
ugt2b5
|
UDP glucuronosyltransferase 2 family, polypeptide B5 |
| chr12_+_31673588 | 2.39 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr4_-_12766418 | 2.39 |
ENSDART00000024312
|
dera
|
deoxyribose-phosphate aldolase (putative) |
| chr10_+_21867307 | 2.37 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr4_-_34480599 | 2.26 |
ENSDART00000159602
ENSDART00000182248 |
si:dkeyp-51g9.4
si:ch211-64i20.3
|
si:dkeyp-51g9.4 si:ch211-64i20.3 |
| chr7_+_40205394 | 2.25 |
ENSDART00000173742
ENSDART00000148390 |
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr18_+_3530769 | 2.18 |
ENSDART00000163469
|
si:ch73-338a16.3
|
si:ch73-338a16.3 |
| chr7_-_4296771 | 2.17 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr8_+_50742975 | 2.12 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr10_+_2981090 | 2.08 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr12_+_47044707 | 2.08 |
ENSDART00000186506
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr15_-_33964897 | 2.07 |
ENSDART00000172075
ENSDART00000158126 ENSDART00000160456 |
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr5_+_28858345 | 2.05 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr21_+_10076203 | 2.03 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr9_-_1459985 | 2.02 |
ENSDART00000124777
|
osbpl6
|
oxysterol binding protein-like 6 |
| chr5_-_65158203 | 2.01 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr1_-_19502322 | 2.00 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr5_+_4054704 | 1.97 |
ENSDART00000140537
|
dhrs11a
|
dehydrogenase/reductase (SDR family) member 11a |
| chr2_+_49417900 | 1.96 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr5_+_9428876 | 1.95 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr6_+_40723554 | 1.91 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr22_-_36770649 | 1.89 |
ENSDART00000169442
|
acy1
|
aminoacylase 1 |
| chr10_-_41664427 | 1.86 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr25_+_4091067 | 1.80 |
ENSDART00000104926
|
eps8l2
|
EPS8 like 2 |
| chr19_-_31042570 | 1.77 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr11_-_2270069 | 1.74 |
ENSDART00000189005
|
znf740a
|
zinc finger protein 740a |
| chr12_+_5209822 | 1.73 |
ENSDART00000152610
|
SLC35G1
|
si:ch211-197g18.2 |
| chr6_-_47953829 | 1.72 |
ENSDART00000157264
|
si:dkey-166d12.2
|
si:dkey-166d12.2 |
| chr6_-_59563597 | 1.72 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr5_+_29851433 | 1.71 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr12_+_2648043 | 1.70 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr16_+_11603096 | 1.68 |
ENSDART00000146286
|
si:dkey-11o1.3
|
si:dkey-11o1.3 |
| chr8_+_1187928 | 1.66 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr17_-_34952562 | 1.66 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr16_+_11623956 | 1.65 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr8_+_999421 | 1.62 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr19_-_22387141 | 1.61 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr3_+_26734162 | 1.60 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr1_-_9134045 | 1.59 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr22_-_23625351 | 1.58 |
ENSDART00000192588
ENSDART00000163423 |
cfhl4
|
complement factor H like 4 |
| chr15_+_14109652 | 1.58 |
ENSDART00000156370
|
clca1
|
chloride channel accessory 1 |
| chr21_+_38002879 | 1.57 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr21_+_39100289 | 1.56 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr24_-_23675446 | 1.56 |
ENSDART00000066644
|
hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr15_+_29283981 | 1.55 |
ENSDART00000099895
|
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr3_+_49097775 | 1.55 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr15_-_17868870 | 1.53 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr5_+_26795773 | 1.52 |
ENSDART00000145631
|
tcn2
|
transcobalamin II |
| chr25_+_19238175 | 1.51 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr3_+_49521106 | 1.50 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr25_+_7670683 | 1.50 |
ENSDART00000040275
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr13_-_42579437 | 1.49 |
ENSDART00000135172
|
fam83ha
|
family with sequence similarity 83, member Ha |
| chr19_-_22568203 | 1.48 |
ENSDART00000182888
|
pleca
|
plectin a |
| chr6_-_56103718 | 1.46 |
ENSDART00000191002
|
gcnt7
|
glucosaminyl (N-acetyl) transferase family member 7 |
| chr8_+_7144066 | 1.43 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr25_+_16098620 | 1.43 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr2_-_5475910 | 1.39 |
ENSDART00000100954
ENSDART00000172143 ENSDART00000132496 |
proca
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr13_-_40726865 | 1.37 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr21_+_21803363 | 1.34 |
ENSDART00000081184
|
neu3.3
|
sialidase 3 (membrane sialidase), tandem duplicate 3 |
| chr21_+_21803596 | 1.32 |
ENSDART00000084600
ENSDART00000184899 |
neu3.3
|
sialidase 3 (membrane sialidase), tandem duplicate 3 |
| chr5_+_13385837 | 1.30 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr1_-_21409877 | 1.30 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr11_-_40647190 | 1.27 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr21_-_3613702 | 1.24 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr18_-_25905574 | 1.19 |
ENSDART00000143899
ENSDART00000163369 |
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr12_-_20373058 | 1.18 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr14_+_8504410 | 1.16 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr18_+_9382847 | 1.16 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr4_+_34119824 | 1.15 |
ENSDART00000165637
|
si:ch211-223g7.6
|
si:ch211-223g7.6 |
| chr24_-_31123365 | 1.13 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr23_+_26491931 | 1.12 |
ENSDART00000190124
ENSDART00000132905 |
si:ch73-206d17.1
|
si:ch73-206d17.1 |
| chr16_-_18702249 | 1.06 |
ENSDART00000191595
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr6_+_13606410 | 1.05 |
ENSDART00000104716
|
asic4b
|
acid-sensing (proton-gated) ion channel family member 4b |
| chr9_-_34269066 | 1.04 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr9_+_30720048 | 1.04 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr5_+_9348284 | 1.03 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr23_-_24394719 | 1.03 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr5_-_29488245 | 1.01 |
ENSDART00000047719
ENSDART00000141154 ENSDART00000171165 |
cacna1ba
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, a |
| chr2_-_5466708 | 1.01 |
ENSDART00000136682
|
proca
|
protein C (inactivator of coagulation factors Va and VIIIa), a |
| chr13_-_23666579 | 0.98 |
ENSDART00000192457
ENSDART00000179777 |
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr4_-_56954002 | 0.98 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr3_+_12718100 | 0.95 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr17_-_44440832 | 0.94 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr17_-_47305034 | 0.93 |
ENSDART00000150116
|
alk
|
ALK receptor tyrosine kinase |
| chr15_+_40079468 | 0.93 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr24_-_3304664 | 0.93 |
ENSDART00000183168
|
sugct
|
succinyl-CoA:glutarate-CoA transferase |
| chr6_+_20647155 | 0.92 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr17_+_1496107 | 0.92 |
ENSDART00000187804
|
LO018430.1
|
|
| chr13_+_2394264 | 0.91 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr3_+_15505275 | 0.90 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr20_+_6543674 | 0.90 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr6_-_18250857 | 0.87 |
ENSDART00000159486
|
anapc11
|
APC11 anaphase promoting complex subunit 11 homolog (yeast) |
| chr3_+_40170216 | 0.85 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr11_-_27962757 | 0.84 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr9_+_29994439 | 0.83 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr22_-_23781083 | 0.83 |
ENSDART00000166563
ENSDART00000170458 ENSDART00000166158 ENSDART00000171246 |
cfhl3
|
complement factor H like 3 |
| chr8_-_18535822 | 0.82 |
ENSDART00000100558
|
nexn
|
nexilin (F actin binding protein) |
| chr20_-_25436389 | 0.82 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr17_+_15033822 | 0.79 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr3_-_34084387 | 0.78 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr21_+_37442458 | 0.77 |
ENSDART00000180161
ENSDART00000186950 |
cox7b
|
cytochrome c oxidase subunit VIIb |
| chr24_-_20016817 | 0.77 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr13_+_48617974 | 0.74 |
ENSDART00000168596
|
si:ch1073-268j14.1
|
si:ch1073-268j14.1 |
| chr3_-_18047150 | 0.73 |
ENSDART00000020376
|
hsd17b1
|
hydroxysteroid (17-beta) dehydrogenase 1 |
| chr8_-_47800754 | 0.71 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr14_+_740032 | 0.70 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr20_+_10544100 | 0.68 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr18_-_5111449 | 0.68 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr21_+_43669943 | 0.68 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr7_-_29625509 | 0.67 |
ENSDART00000173723
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr14_-_41556533 | 0.61 |
ENSDART00000074401
|
itga6l
|
integrin, alpha 6, like |
| chr10_+_20108557 | 0.59 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr25_+_23280220 | 0.56 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr22_-_23706771 | 0.53 |
ENSDART00000159771
|
cfhl1
|
complement factor H like 1 |
| chr14_+_23874062 | 0.51 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr7_-_61901695 | 0.49 |
ENSDART00000110328
|
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr2_-_44746723 | 0.48 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium chain family member 3 |
| chr5_+_45322734 | 0.47 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr2_-_43635777 | 0.45 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr2_-_48396061 | 0.43 |
ENSDART00000161657
|
si:ch211-195j11.30
|
si:ch211-195j11.30 |
| chr11_-_19501439 | 0.39 |
ENSDART00000158328
ENSDART00000103983 |
slc2a9l1
|
solute carrier family 2 (facilitated glucose transporter), member 9-like 1 |
| chr6_-_49078702 | 0.39 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr17_+_30591287 | 0.38 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr6_-_37744430 | 0.37 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr3_-_6191950 | 0.34 |
ENSDART00000161931
|
si:ch73-144l3.1
|
si:ch73-144l3.1 |
| chr4_-_32642681 | 0.34 |
ENSDART00000188695
ENSDART00000193974 |
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr20_+_18740799 | 0.34 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr18_+_13182528 | 0.33 |
ENSDART00000166298
|
zgc:56622
|
zgc:56622 |
| chr23_+_25856541 | 0.33 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr20_-_7293837 | 0.32 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr22_-_36750589 | 0.32 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
| chr21_-_38853737 | 0.30 |
ENSDART00000184100
|
tlr22
|
toll-like receptor 22 |
| chr16_+_4138331 | 0.30 |
ENSDART00000192403
ENSDART00000193036 |
mtf1
|
metal-regulatory transcription factor 1 |
| chr8_-_13013123 | 0.29 |
ENSDART00000147802
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr2_-_24402341 | 0.29 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr22_+_12770877 | 0.28 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr9_-_43538328 | 0.26 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr13_+_38430466 | 0.25 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr24_+_14713776 | 0.25 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr25_+_16194979 | 0.24 |
ENSDART00000185592
ENSDART00000158582 ENSDART00000161109 ENSDART00000139013 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr14_-_11507211 | 0.24 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr11_-_13341483 | 0.23 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr15_+_9297340 | 0.23 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr4_+_2620751 | 0.16 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr6_+_28140816 | 0.16 |
ENSDART00000137907
|
inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr2_-_10943093 | 0.16 |
ENSDART00000148999
|
ssbp3a
|
single stranded DNA binding protein 3a |
| chr15_-_17869115 | 0.15 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr11_-_12801157 | 0.13 |
ENSDART00000103449
|
txlng
|
taxilin gamma |
| chr4_+_45484774 | 0.11 |
ENSDART00000150573
|
si:dkey-256i11.6
|
si:dkey-256i11.6 |
| chr11_-_12800945 | 0.11 |
ENSDART00000191178
|
txlng
|
taxilin gamma |
| chr21_+_11535304 | 0.08 |
ENSDART00000158901
ENSDART00000081600 |
cd8b
|
cd8 beta |
Network of associatons between targets according to the STRING database.
First level regulatory network of hnf1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 1.2 | 3.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.6 | 8.0 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.5 | 1.5 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.5 | 11.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 2.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.5 | 5.0 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.4 | 2.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.4 | 3.3 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.4 | 5.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 10.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.3 | 29.4 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.3 | 1.9 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.3 | 2.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 4.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.8 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 2.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.9 | GO:0031179 | peptide modification(GO:0031179) |
| 0.2 | 1.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 1.6 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 0.7 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.2 | 2.7 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.2 | 2.4 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.2 | 2.9 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.2 | 18.6 | GO:0009410 | response to xenobiotic stimulus(GO:0009410) |
| 0.2 | 2.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.2 | 3.0 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 3.1 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.1 | 2.8 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.2 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.7 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 2.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 2.0 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.3 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 3.0 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.1 | 0.3 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.1 | 1.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.9 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 2.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 1.0 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 2.5 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 2.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 1.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.0 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 3.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 1.9 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.9 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 4.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.0 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.6 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 1.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 2.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 1.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 1.2 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 3.0 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 3.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.8 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 1.6 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 18.0 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 5.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 2.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.2 | 0.7 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 2.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 5.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.6 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 3.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 6.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 7.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 48.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 7.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 5.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 13.8 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.7 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 5.0 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.8 | 3.3 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.8 | 2.5 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.7 | 13.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.7 | 4.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.6 | 32.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.5 | 3.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.5 | 2.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.5 | 6.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.5 | 1.4 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.4 | 1.7 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.4 | 1.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.4 | 10.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 1.5 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.3 | 0.9 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.3 | 29.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.7 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.2 | 2.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 6.1 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.9 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.7 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 2.9 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 2.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.1 | 2.1 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 10.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 2.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 3.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.9 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 8.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 1.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 3.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 4.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.4 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 3.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.5 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 1.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 2.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 16.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 4.7 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 8.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.5 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 3.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 2.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 5.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 10.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 1.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 3.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 3.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 5.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 3.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 1.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 2.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 5.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 3.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 3.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.9 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 5.6 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |