Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
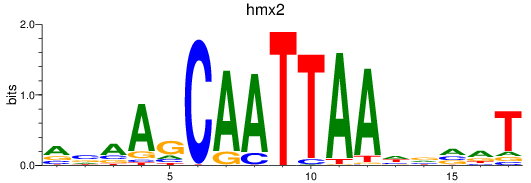
Results for hmx2_hmx3a
Z-value: 1.15
Transcription factors associated with hmx2_hmx3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx2
|
ENSDARG00000070954 | H6 family homeobox 2 |
|
hmx2
|
ENSDARG00000115364 | H6 family homeobox 2 |
|
hmx3a
|
ENSDARG00000070955 | H6 family homeobox 3a |
|
hmx3a
|
ENSDARG00000115051 | H6 family homeobox 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx2 | dr11_v1_chr17_-_21784152_21784152 | -0.63 | 3.8e-03 | Click! |
| hmx3a | dr11_v1_chr17_-_21793113_21793113 | -0.52 | 2.4e-02 | Click! |
Activity profile of hmx2_hmx3a motif
Sorted Z-values of hmx2_hmx3a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_60721342 | 7.42 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr4_-_9909371 | 3.47 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr10_-_1961930 | 3.30 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr16_+_54209504 | 2.94 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr24_+_38301080 | 2.83 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr3_-_27647845 | 2.28 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr2_-_47431205 | 1.92 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr8_-_22965916 | 1.92 |
ENSDART00000143791
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr4_+_25607101 | 1.56 |
ENSDART00000133929
|
acot14
|
acyl-CoA thioesterase 14 |
| chr11_-_41966854 | 1.49 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr2_+_42005475 | 1.49 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr22_-_9906790 | 1.48 |
ENSDART00000105936
|
si:dkey-253d23.8
|
si:dkey-253d23.8 |
| chr16_+_40043673 | 1.45 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr11_+_18157260 | 1.38 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr21_+_8239544 | 1.34 |
ENSDART00000122773
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr1_-_25438737 | 1.33 |
ENSDART00000134470
|
fhdc1
|
FH2 domain containing 1 |
| chr12_+_13118540 | 1.32 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr4_+_22297839 | 1.29 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr23_+_32029304 | 1.27 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr11_+_18130300 | 1.25 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr4_+_60934218 | 1.24 |
ENSDART00000150675
|
si:dkey-82i20.2
|
si:dkey-82i20.2 |
| chr4_-_50759477 | 1.21 |
ENSDART00000150246
|
si:ch211-245n8.4
|
si:ch211-245n8.4 |
| chr16_-_28268201 | 1.20 |
ENSDART00000121671
ENSDART00000141911 |
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr3_+_26244353 | 1.17 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr17_+_16429826 | 1.15 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr20_+_25225112 | 1.15 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr4_-_4592287 | 1.15 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr21_+_3796196 | 1.14 |
ENSDART00000146754
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr2_-_24962002 | 1.12 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr4_+_25607743 | 1.12 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr20_+_48129759 | 1.11 |
ENSDART00000148494
|
tram2
|
translocation associated membrane protein 2 |
| chr21_-_23307653 | 1.10 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr4_-_18741942 | 1.10 |
ENSDART00000145747
ENSDART00000066980 ENSDART00000186518 |
atxn10
|
ataxin 10 |
| chr8_-_22538588 | 1.08 |
ENSDART00000144041
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr8_+_28358161 | 1.07 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr1_-_27014872 | 1.07 |
ENSDART00000147414
ENSDART00000134032 ENSDART00000192087 ENSDART00000189111 ENSDART00000187348 ENSDART00000187248 |
cntln
|
centlein, centrosomal protein |
| chr20_-_23171430 | 1.05 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr5_+_32835219 | 1.05 |
ENSDART00000140832
ENSDART00000186055 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr5_-_25723079 | 1.03 |
ENSDART00000014013
|
gda
|
guanine deaminase |
| chr17_-_27223965 | 1.03 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr20_+_4221978 | 1.01 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr23_+_32028574 | 1.01 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr7_+_23940933 | 1.00 |
ENSDART00000173628
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr9_+_52621487 | 0.99 |
ENSDART00000166266
|
si:ch211-241j8.2
|
si:ch211-241j8.2 |
| chr22_+_2510828 | 0.99 |
ENSDART00000115348
ENSDART00000145611 |
zgc:173726
|
zgc:173726 |
| chr3_+_28502419 | 0.99 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr22_+_2511045 | 0.99 |
ENSDART00000106425
|
zgc:173726
|
zgc:173726 |
| chr18_+_36582815 | 0.99 |
ENSDART00000059311
|
siae
|
sialic acid acetylesterase |
| chr18_-_25568994 | 0.98 |
ENSDART00000133029
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr4_+_50310886 | 0.98 |
ENSDART00000182141
|
znf1066
|
zinc finger protein 1066 |
| chr12_+_19188542 | 0.98 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr20_+_98179 | 0.97 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr13_-_31008275 | 0.96 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr9_-_14273652 | 0.95 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr22_-_4649238 | 0.95 |
ENSDART00000125302
|
fbn2b
|
fibrillin 2b |
| chr24_-_16905018 | 0.95 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr24_+_14541013 | 0.95 |
ENSDART00000066721
|
ngdn
|
neuroguidin, EIF4E binding protein |
| chr13_-_40316367 | 0.94 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr9_+_48761455 | 0.93 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr4_-_48634516 | 0.93 |
ENSDART00000129731
|
znf1063
|
zinc finger protein 1063 |
| chr24_-_3477103 | 0.92 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr21_-_37973819 | 0.92 |
ENSDART00000133405
|
ripply1
|
ripply transcriptional repressor 1 |
| chr6_-_26080384 | 0.91 |
ENSDART00000157181
ENSDART00000154568 |
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr8_+_45381298 | 0.90 |
ENSDART00000149451
ENSDART00000110364 |
tti2
|
TELO2 interacting protein 2 |
| chr25_+_34247353 | 0.89 |
ENSDART00000148914
|
bnip2
|
BCL2 interacting protein 2 |
| chr5_+_18014931 | 0.88 |
ENSDART00000142562
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr24_+_5789790 | 0.88 |
ENSDART00000189600
|
BX470065.1
|
|
| chr19_-_22328154 | 0.88 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr16_+_28994709 | 0.86 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr15_-_41689981 | 0.84 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr24_-_5911973 | 0.84 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr4_-_12930086 | 0.84 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr7_+_39399747 | 0.83 |
ENSDART00000147037
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr3_-_16719244 | 0.81 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr21_-_22117085 | 0.81 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr15_-_23529945 | 0.81 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr11_-_34577034 | 0.80 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr16_-_14397003 | 0.79 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr2_+_15128418 | 0.78 |
ENSDART00000141921
|
arhgap29b
|
Rho GTPase activating protein 29b |
| chr16_-_45854882 | 0.78 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr16_-_9830451 | 0.77 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr4_+_40302769 | 0.77 |
ENSDART00000137892
|
znf993
|
zinc finger protein 993 |
| chr13_+_1131748 | 0.77 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr17_-_16422654 | 0.77 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr6_+_27418541 | 0.76 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr7_+_2455344 | 0.76 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr4_+_37092809 | 0.76 |
ENSDART00000174458
|
BX571760.1
|
|
| chr24_-_10014512 | 0.74 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr12_+_1609563 | 0.74 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr14_+_52481288 | 0.74 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr21_+_18907102 | 0.74 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr12_+_31537604 | 0.73 |
ENSDART00000153340
|
si:ch73-205h11.1
|
si:ch73-205h11.1 |
| chr14_-_17262082 | 0.73 |
ENSDART00000173944
ENSDART00000172317 ENSDART00000171064 |
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr21_+_15601100 | 0.72 |
ENSDART00000180558
ENSDART00000145454 |
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr4_+_41159678 | 0.72 |
ENSDART00000131561
|
znf1062
|
zinc finger protein 1062 |
| chr17_+_25197180 | 0.72 |
ENSDART00000148775
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr23_+_9353552 | 0.72 |
ENSDART00000163298
|
BX511246.1
|
|
| chr4_+_33423119 | 0.71 |
ENSDART00000189413
|
znf1065
|
zinc finger protein 1065 |
| chr16_-_7793457 | 0.71 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr3_-_36458143 | 0.71 |
ENSDART00000168733
ENSDART00000167741 ENSDART00000167143 |
glyr1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr15_-_18361475 | 0.70 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr6_-_30683637 | 0.70 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr3_-_58644920 | 0.70 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr13_+_41917606 | 0.69 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr20_-_15090862 | 0.69 |
ENSDART00000063892
ENSDART00000122592 |
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr3_-_26017592 | 0.69 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_+_32410746 | 0.69 |
ENSDART00000025496
|
rras
|
RAS related |
| chr14_+_31657412 | 0.68 |
ENSDART00000105767
|
fhl1a
|
four and a half LIM domains 1a |
| chr8_+_21437908 | 0.68 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr3_-_40965596 | 0.68 |
ENSDART00000137188
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr21_+_6394929 | 0.67 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr7_+_15736230 | 0.67 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr7_+_29163762 | 0.67 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr5_-_14326959 | 0.67 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr12_-_8504278 | 0.67 |
ENSDART00000135865
|
egr2b
|
early growth response 2b |
| chr11_+_14321113 | 0.66 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr22_+_1751640 | 0.66 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr21_+_39941875 | 0.66 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr3_-_26806032 | 0.66 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr14_-_6285555 | 0.65 |
ENSDART00000182280
ENSDART00000147184 |
elp1
|
elongator complex protein 1 |
| chr20_+_25445826 | 0.65 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr13_-_16066997 | 0.63 |
ENSDART00000184790
|
SPATA48
|
spermatogenesis associated 48 |
| chr13_+_7442023 | 0.63 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr13_+_2894536 | 0.63 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr19_-_5699703 | 0.63 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr2_+_26655744 | 0.62 |
ENSDART00000174928
|
asph
|
aspartate beta-hydroxylase |
| chr24_+_14527935 | 0.62 |
ENSDART00000134846
|
si:dkeyp-73g8.5
|
si:dkeyp-73g8.5 |
| chr19_-_46037835 | 0.61 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr6_+_43234213 | 0.61 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr3_-_40965328 | 0.61 |
ENSDART00000102393
|
cyp3c2
|
cytochrome P450, family 3, subfamily c, polypeptide 2 |
| chr6_+_29410986 | 0.61 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr11_+_31324335 | 0.61 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr9_+_38457806 | 0.60 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr15_-_20839763 | 0.60 |
ENSDART00000141746
ENSDART00000182369 |
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr1_+_25650917 | 0.60 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr16_-_31790285 | 0.60 |
ENSDART00000184655
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr2_-_44777592 | 0.60 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr5_+_26847190 | 0.60 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr15_+_627619 | 0.59 |
ENSDART00000157207
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr2_+_44348473 | 0.59 |
ENSDART00000155166
ENSDART00000098146 |
zgc:152670
|
zgc:152670 |
| chr17_+_50701748 | 0.59 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr23_+_21424747 | 0.59 |
ENSDART00000135440
ENSDART00000104214 |
tas1r1
|
taste receptor, type 1, member 1 |
| chr6_+_3717613 | 0.58 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr24_+_16905188 | 0.58 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr21_-_17296789 | 0.58 |
ENSDART00000192180
|
gfi1b
|
growth factor independent 1B transcription repressor |
| chr15_+_2856696 | 0.58 |
ENSDART00000163434
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr16_+_14707960 | 0.58 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr20_-_34670236 | 0.58 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr22_+_2751887 | 0.57 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr13_+_18321140 | 0.57 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr14_+_23709134 | 0.57 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr4_+_42081504 | 0.57 |
ENSDART00000162361
|
znf1060
|
zinc finger protein 1060 |
| chr3_+_12725343 | 0.57 |
ENSDART00000188583
|
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr9_+_24106469 | 0.56 |
ENSDART00000039295
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr9_+_30720048 | 0.56 |
ENSDART00000146115
|
klf12b
|
Kruppel-like factor 12b |
| chr11_-_41853874 | 0.56 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr24_+_5912898 | 0.55 |
ENSDART00000132686
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr7_-_4461104 | 0.55 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr2_-_40890004 | 0.55 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr17_+_24318753 | 0.55 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr22_+_2769236 | 0.55 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr23_+_2917392 | 0.55 |
ENSDART00000150019
|
DHX35
|
zgc:158828 |
| chr17_+_8542203 | 0.55 |
ENSDART00000158873
|
CU462878.2
|
|
| chr1_+_49682312 | 0.54 |
ENSDART00000136747
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr20_-_2641233 | 0.54 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr16_+_5901835 | 0.54 |
ENSDART00000060519
|
ulk4
|
unc-51 like kinase 4 |
| chr1_-_2449395 | 0.54 |
ENSDART00000103785
|
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr1_-_52461322 | 0.54 |
ENSDART00000083836
|
si:ch211-217k17.7
|
si:ch211-217k17.7 |
| chr20_-_16972351 | 0.53 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr23_-_36303216 | 0.53 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr8_-_944055 | 0.53 |
ENSDART00000092773
|
mrps27
|
mitochondrial ribosomal protein S27 |
| chr15_+_857148 | 0.53 |
ENSDART00000156949
|
si:dkey-7i4.13
|
si:dkey-7i4.13 |
| chr3_-_26244256 | 0.53 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr11_+_31323746 | 0.53 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr8_+_28259347 | 0.53 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr19_-_17303500 | 0.53 |
ENSDART00000162355
|
sf3a3
|
splicing factor 3a, subunit 3 |
| chr1_-_2448592 | 0.53 |
ENSDART00000190207
|
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr25_+_34247107 | 0.53 |
ENSDART00000148507
|
bnip2
|
BCL2 interacting protein 2 |
| chr5_+_30392148 | 0.52 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr19_-_19379084 | 0.52 |
ENSDART00000165206
|
smarcc1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
| chr11_-_20988238 | 0.52 |
ENSDART00000155238
|
taf4a
|
TAF4A RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr22_-_26100282 | 0.52 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr19_+_43359075 | 0.52 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr19_-_1002959 | 0.52 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr4_-_56151174 | 0.51 |
ENSDART00000125904
|
znf986
|
zinc finger protein 986 |
| chr22_+_21305682 | 0.51 |
ENSDART00000023521
|
odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr6_-_39270851 | 0.51 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr23_-_16980213 | 0.50 |
ENSDART00000046889
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr4_+_39141908 | 0.50 |
ENSDART00000189450
|
CR407594.1
|
|
| chr20_-_33566640 | 0.50 |
ENSDART00000159729
|
si:dkey-65b13.9
|
si:dkey-65b13.9 |
| chr3_+_54761569 | 0.50 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr7_-_26270014 | 0.50 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr22_-_26005894 | 0.50 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr22_+_20172018 | 0.50 |
ENSDART00000188104
|
hmg20b
|
high mobility group 20B |
| chr7_-_25697285 | 0.50 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr6_-_19341184 | 0.50 |
ENSDART00000168236
ENSDART00000167674 |
mif4gda
|
MIF4G domain containing a |
| chr19_-_868187 | 0.49 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr14_-_9085349 | 0.49 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| chr6_+_3716666 | 0.49 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr15_+_11814969 | 0.49 |
ENSDART00000127248
|
FO704748.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx2_hmx3a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 1.1 | 3.3 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.7 | 3.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.4 | 0.4 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.4 | 1.2 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.3 | 1.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 1.0 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.3 | 1.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 1.0 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.3 | 1.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 1.5 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.2 | 0.9 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.2 | 0.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 0.6 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 1.0 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.2 | 0.8 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.2 | 0.8 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 0.9 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 2.3 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.2 | 0.5 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.2 | 1.1 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.2 | 1.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 1.7 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.6 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:2000726 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 1.9 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.1 | 0.9 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.1 | 0.4 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.0 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 1.8 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.7 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.5 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.6 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.5 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.1 | 0.7 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 1.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 1.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 1.2 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.2 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.5 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.3 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.0 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.3 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.7 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.5 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.3 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.4 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 1.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.4 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.1 | 0.4 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.1 | 0.3 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.4 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 1.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.7 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.3 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.4 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.9 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.3 | GO:0035767 | larval development(GO:0002164) larval heart development(GO:0007508) endothelial cell chemotaxis(GO:0035767) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.0 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.2 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.3 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.8 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.1 | 0.2 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.2 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 0.2 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.3 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.7 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.2 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.7 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.7 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.7 | GO:0048932 | myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.2 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.5 | GO:1901888 | regulation of cell junction assembly(GO:1901888) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 1.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.3 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 1.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 1.5 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.1 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.3 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.6 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 2.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.5 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.3 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.6 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.1 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.0 | 1.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.8 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.8 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) negative regulation of neural precursor cell proliferation(GO:2000178) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.0 | GO:0072679 | thymocyte migration(GO:0072679) |
| 0.0 | 0.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.2 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 4.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.1 | GO:0003315 | heart rudiment morphogenesis(GO:0003314) heart rudiment formation(GO:0003315) |
| 0.0 | 0.0 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 2.6 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.0 | GO:0061469 | type B pancreatic cell proliferation(GO:0044342) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 1.8 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.1 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.3 | 1.3 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 0.6 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 2.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 1.0 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 0.8 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 1.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.6 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.8 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.6 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.0 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 1.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.3 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.6 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.5 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.4 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.4 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.9 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.3 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.4 | 1.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.3 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.3 | 1.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 0.9 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.3 | 0.8 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.3 | 0.8 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 3.2 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 0.9 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.2 | 2.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 0.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 0.6 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.8 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 1.0 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 1.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 0.6 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.2 | 1.7 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.4 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.9 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.4 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.4 | GO:0032405 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.7 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.7 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 3.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 1.7 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 2.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 1.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.3 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.8 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.6 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.6 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 2.6 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.7 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 1.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0000996 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 2.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 1.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 2.3 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0071916 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 29.5 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0051117 | ATPase binding(GO:0051117) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 3.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.9 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 1.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 0.8 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.1 | 0.8 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.8 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |