Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
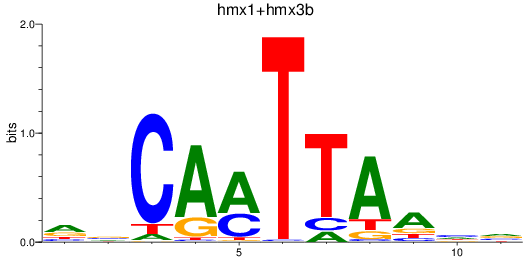
Results for hmx1+hmx3b
Z-value: 0.57
Transcription factors associated with hmx1+hmx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmx1
|
ENSDARG00000095651 | H6 family homeobox 1 |
|
hmx3b
|
ENSDARG00000096716 | H6 family homeobox 3b |
|
hmx1
|
ENSDARG00000109725 | H6 family homeobox 1 |
|
hmx1
|
ENSDARG00000111765 | H6 family homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmx3b | dr11_v1_chr12_-_49154934_49154934 | 0.56 | 1.2e-02 | Click! |
| hmx1 | dr11_v1_chr1_-_40914752_40914752 | -0.31 | 1.9e-01 | Click! |
Activity profile of hmx1+hmx3b motif
Sorted Z-values of hmx1+hmx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_30244356 | 2.85 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr25_-_10503043 | 1.48 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr4_-_72609735 | 1.22 |
ENSDART00000174299
ENSDART00000159227 |
si:cabz01054394.6
|
si:cabz01054394.6 |
| chr14_-_33454595 | 1.20 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr7_+_66565930 | 1.19 |
ENSDART00000154597
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr8_+_24861264 | 1.17 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr1_-_22512063 | 1.15 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr21_+_21906671 | 1.15 |
ENSDART00000016916
|
gria4b
|
glutamate receptor, ionotropic, AMPA 4b |
| chr3_+_22327738 | 1.10 |
ENSDART00000055675
|
gh1
|
growth hormone 1 |
| chr6_-_40697585 | 1.10 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr5_-_42272517 | 1.03 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr25_+_3326885 | 1.00 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr11_+_29770966 | 0.97 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr12_+_24342303 | 0.97 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr12_+_2446837 | 0.96 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr24_-_27400017 | 0.95 |
ENSDART00000145829
|
ccl34b.1
|
chemokine (C-C motif) ligand 34b, duplicate 1 |
| chr2_-_16380283 | 0.81 |
ENSDART00000149992
|
si:dkey-231j24.3
|
si:dkey-231j24.3 |
| chr9_-_36924388 | 0.81 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr20_-_34750045 | 0.76 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr7_+_32722227 | 0.73 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr21_-_14803366 | 0.73 |
ENSDART00000190872
|
si:dkey-11o18.5
|
si:dkey-11o18.5 |
| chr11_-_37691449 | 0.71 |
ENSDART00000185340
|
zgc:158258
|
zgc:158258 |
| chr8_-_19649617 | 0.64 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr20_-_7128612 | 0.63 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr25_+_3327071 | 0.62 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr20_+_37295006 | 0.62 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr23_-_36857964 | 0.61 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr10_-_33297864 | 0.59 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr10_-_39153959 | 0.58 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr20_+_28861629 | 0.58 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr19_+_9295244 | 0.57 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr22_+_724639 | 0.53 |
ENSDART00000105323
|
zgc:162255
|
zgc:162255 |
| chr22_-_7129631 | 0.52 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr15_+_1134870 | 0.52 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr1_+_34696503 | 0.49 |
ENSDART00000186106
|
CR339054.2
|
|
| chr19_-_6239248 | 0.47 |
ENSDART00000014127
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr4_-_837768 | 0.46 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr2_+_54798689 | 0.46 |
ENSDART00000183426
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr21_-_14966718 | 0.44 |
ENSDART00000151200
|
mmp17a
|
matrix metallopeptidase 17a |
| chr10_-_39154594 | 0.44 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr1_+_52929185 | 0.44 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr18_+_11970987 | 0.43 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr13_-_36911118 | 0.42 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr8_-_46486009 | 0.39 |
ENSDART00000140431
|
sult1st9
|
sulfotransferase family 1, cytosolic sulfotransferase 9 |
| chr13_+_23176330 | 0.39 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr5_+_24287927 | 0.36 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr22_-_6988102 | 0.35 |
ENSDART00000185618
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr4_-_4751981 | 0.33 |
ENSDART00000147436
ENSDART00000092984 ENSDART00000158466 |
creb3l2
|
cAMP responsive element binding protein 3-like 2 |
| chr23_+_38953545 | 0.33 |
ENSDART00000184621
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr8_+_22472584 | 0.32 |
ENSDART00000138303
|
si:dkey-23c22.9
|
si:dkey-23c22.9 |
| chr3_+_56366395 | 0.32 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr5_-_69620722 | 0.32 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr20_-_5052786 | 0.32 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr18_-_5595546 | 0.31 |
ENSDART00000191825
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr13_-_4018888 | 0.31 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr11_-_7261717 | 0.29 |
ENSDART00000128959
|
zgc:113223
|
zgc:113223 |
| chr1_-_23596391 | 0.29 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr20_+_28861435 | 0.27 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr5_+_19165949 | 0.25 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr9_-_25425381 | 0.25 |
ENSDART00000129522
|
acvr2aa
|
activin A receptor type 2Aa |
| chr18_-_33080454 | 0.24 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr10_+_43039947 | 0.19 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr6_+_4528631 | 0.19 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr21_+_21702623 | 0.19 |
ENSDART00000136840
|
or125-2
|
odorant receptor, family E, subfamily 125, member 2 |
| chr5_+_11290851 | 0.18 |
ENSDART00000180408
|
CABZ01077124.1
|
|
| chr8_+_7097740 | 0.18 |
ENSDART00000159670
|
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr15_-_1822548 | 0.17 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr5_-_25645593 | 0.17 |
ENSDART00000044471
|
tmc2b
|
transmembrane channel-like 2b |
| chrM_+_8130 | 0.16 |
ENSDART00000093609
|
mt-co2
|
cytochrome c oxidase II, mitochondrial |
| chr19_+_11217279 | 0.16 |
ENSDART00000181859
|
si:ch73-109i22.2
|
si:ch73-109i22.2 |
| chr24_+_26134209 | 0.13 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr22_+_14117078 | 0.13 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr5_+_37032038 | 0.13 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr6_+_11681011 | 0.13 |
ENSDART00000151447
ENSDART00000151618 |
calcrlb
|
calcitonin receptor-like b |
| chr19_+_43604256 | 0.11 |
ENSDART00000151080
ENSDART00000110305 |
si:ch211-199g17.9
|
si:ch211-199g17.9 |
| chr17_+_50076501 | 0.10 |
ENSDART00000156303
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr22_-_4649238 | 0.10 |
ENSDART00000125302
|
fbn2b
|
fibrillin 2b |
| chr16_-_15988320 | 0.08 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr5_+_32009542 | 0.07 |
ENSDART00000182025
ENSDART00000179879 |
scai
|
suppressor of cancer cell invasion |
| chr22_-_20695237 | 0.07 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr16_-_35427060 | 0.07 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr13_+_30169681 | 0.07 |
ENSDART00000138326
|
pald1b
|
phosphatase domain containing, paladin 1b |
| chr7_-_56766973 | 0.06 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr13_-_37109987 | 0.05 |
ENSDART00000136750
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chrM_+_4993 | 0.04 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr17_+_6563307 | 0.04 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr14_-_9128919 | 0.04 |
ENSDART00000108641
|
sh2d1ab
|
SH2 domain containing 1A duplicate b |
| chr6_+_40951227 | 0.03 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmx1+hmx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0015824 | proline transport(GO:0015824) |
| 0.2 | 1.1 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 1.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.5 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.2 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.6 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.9 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.2 | 1.6 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.5 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.4 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 1.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 2.2 | GO:0015293 | symporter activity(GO:0015293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |