Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
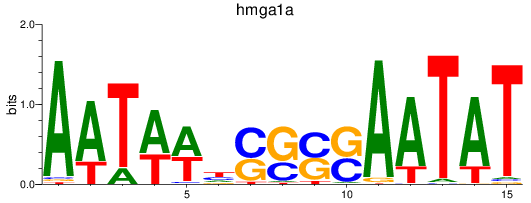
Results for hmga1a
Z-value: 0.70
Transcription factors associated with hmga1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmga1a
|
ENSDARG00000028335 | high mobility group AT-hook 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hmga1a | dr11_v1_chr23_-_3759692_3759692 | 0.96 | 1.6e-10 | Click! |
Activity profile of hmga1a motif
Sorted Z-values of hmga1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_20195350 | 3.31 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr7_-_64971839 | 2.02 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr16_+_1383914 | 1.96 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr14_-_38827442 | 1.80 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr14_-_38828057 | 1.46 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_29808699 | 1.33 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr1_+_47335038 | 1.18 |
ENSDART00000188153
|
bcl9
|
B cell CLL/lymphoma 9 |
| chr13_+_41917606 | 1.16 |
ENSDART00000114741
|
polr1b
|
polymerase (RNA) I polypeptide B |
| chr7_+_33130639 | 1.12 |
ENSDART00000142450
ENSDART00000173967 ENSDART00000173832 |
zgc:153219
si:ch211-194p6.7
|
zgc:153219 si:ch211-194p6.7 |
| chr5_+_32831561 | 1.10 |
ENSDART00000169358
ENSDART00000192078 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr19_+_29808471 | 1.07 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr18_-_35407289 | 1.05 |
ENSDART00000012018
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr17_-_40110782 | 1.04 |
ENSDART00000126929
|
si:dkey-187k19.2
|
si:dkey-187k19.2 |
| chr24_-_15159658 | 1.01 |
ENSDART00000142473
|
rttn
|
rotatin |
| chr24_-_31090948 | 0.99 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr18_+_328689 | 0.94 |
ENSDART00000167841
|
ss18l2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr2_+_25839193 | 0.93 |
ENSDART00000078634
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr20_+_35255403 | 0.90 |
ENSDART00000002773
ENSDART00000137915 |
fam49a
|
family with sequence similarity 49, member A |
| chr1_-_54972170 | 0.88 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr6_-_39518489 | 0.87 |
ENSDART00000185446
|
atf1
|
activating transcription factor 1 |
| chr7_-_33130552 | 0.86 |
ENSDART00000127006
|
mns1
|
meiosis-specific nuclear structural 1 |
| chr10_+_20364009 | 0.84 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr6_-_58764672 | 0.81 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr2_+_32846602 | 0.80 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr3_-_34136368 | 0.80 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr5_-_39805874 | 0.79 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr6_-_58757131 | 0.78 |
ENSDART00000083582
|
soat2
|
sterol O-acyltransferase 2 |
| chr14_-_34513103 | 0.78 |
ENSDART00000136306
|
zgc:194246
|
zgc:194246 |
| chr19_-_20403507 | 0.77 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr22_-_784110 | 0.76 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr16_-_42969598 | 0.75 |
ENSDART00000156011
|
si:ch211-135n15.3
|
si:ch211-135n15.3 |
| chr9_+_25568839 | 0.73 |
ENSDART00000177342
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr18_-_35407695 | 0.72 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr18_-_35407530 | 0.72 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_-_2222707 | 0.69 |
ENSDART00000022179
|
prkag1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr9_-_27738110 | 0.69 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr25_-_13789955 | 0.69 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr4_-_4535189 | 0.67 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr6_+_30533504 | 0.65 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr1_-_54971968 | 0.65 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr23_-_28347039 | 0.65 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr24_-_35534273 | 0.64 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr15_-_33807758 | 0.62 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr9_-_12885201 | 0.62 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr9_-_2892250 | 0.61 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr1_+_31658011 | 0.61 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr3_+_62161184 | 0.59 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr23_+_25354856 | 0.58 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr23_+_38940700 | 0.58 |
ENSDART00000065331
|
sall4
|
spalt-like transcription factor 4 |
| chr7_+_15871156 | 0.57 |
ENSDART00000145946
|
pax6b
|
paired box 6b |
| chr15_+_888704 | 0.54 |
ENSDART00000182796
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr15_+_47440477 | 0.54 |
ENSDART00000002384
|
phox2a
|
paired-like homeobox 2a |
| chr4_+_13568469 | 0.51 |
ENSDART00000171235
ENSDART00000136152 |
calua
|
calumenin a |
| chr23_-_45407631 | 0.51 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr10_+_42733210 | 0.48 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr14_+_26566936 | 0.48 |
ENSDART00000173056
|
arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr8_-_50981175 | 0.48 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr15_-_29387446 | 0.48 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr25_-_34670413 | 0.47 |
ENSDART00000073440
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr7_-_22956889 | 0.45 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr6_-_7123210 | 0.44 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr13_-_41917050 | 0.42 |
ENSDART00000110549
|
cisd1
|
CDGSH iron sulfur domain 1 |
| chr21_+_21743599 | 0.41 |
ENSDART00000101700
|
pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr7_+_19615056 | 0.40 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr6_+_28294113 | 0.38 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_-_22956716 | 0.38 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr16_-_29557338 | 0.38 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr12_+_33919502 | 0.37 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr5_-_20814576 | 0.37 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr19_-_3741602 | 0.36 |
ENSDART00000170301
|
btr22
|
bloodthirsty-related gene family, member 22 |
| chr6_+_22679610 | 0.35 |
ENSDART00000102701
|
zmp:0000000634
|
zmp:0000000634 |
| chr10_+_6121558 | 0.34 |
ENSDART00000166799
ENSDART00000157947 |
tln1
|
talin 1 |
| chr20_+_26349002 | 0.34 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr14_-_15699528 | 0.33 |
ENSDART00000161123
|
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr15_+_17030941 | 0.33 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr25_+_19739665 | 0.32 |
ENSDART00000067353
|
zgc:101783
|
zgc:101783 |
| chr5_+_41476443 | 0.31 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr14_-_33521071 | 0.31 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr20_+_34400715 | 0.30 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr15_+_17030473 | 0.29 |
ENSDART00000129407
|
plin2
|
perilipin 2 |
| chr5_-_38451082 | 0.29 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr9_-_41507712 | 0.29 |
ENSDART00000135821
|
mfsd6b
|
major facilitator superfamily domain containing 6b |
| chr12_-_10409961 | 0.28 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr25_+_15287036 | 0.27 |
ENSDART00000147572
|
hipk3a
|
homeodomain interacting protein kinase 3a |
| chr12_+_46386983 | 0.26 |
ENSDART00000183982
|
BX005305.3
|
Danio rerio legumain (LOC100005356), mRNA. |
| chr25_-_2355107 | 0.26 |
ENSDART00000056121
|
mrps35
|
mitochondrial ribosomal protein S35 |
| chr12_+_46425800 | 0.26 |
ENSDART00000191965
|
BX005305.1
|
|
| chr4_+_17245005 | 0.24 |
ENSDART00000027645
|
casc1
|
cancer susceptibility candidate 1 |
| chr16_-_26232411 | 0.23 |
ENSDART00000139355
|
arhgef1b
|
Rho guanine nucleotide exchange factor (GEF) 1b |
| chr4_+_17245217 | 0.23 |
ENSDART00000184215
ENSDART00000110908 |
casc1
|
cancer susceptibility candidate 1 |
| chr12_+_46404307 | 0.23 |
ENSDART00000185011
|
BX005305.4
|
|
| chr25_-_17910714 | 0.21 |
ENSDART00000191586
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr12_+_46512881 | 0.21 |
ENSDART00000105454
|
CU855711.1
|
|
| chr10_-_26202766 | 0.20 |
ENSDART00000136393
|
fhdc3
|
FH2 domain containing 3 |
| chr24_-_16975960 | 0.20 |
ENSDART00000180687
|
klhl15
|
kelch-like family member 15 |
| chr5_-_1203455 | 0.20 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr12_+_46443477 | 0.20 |
ENSDART00000191873
|
BX005305.1
|
|
| chr8_-_49725430 | 0.18 |
ENSDART00000135675
|
gkap1
|
G kinase anchoring protein 1 |
| chr17_-_21200406 | 0.18 |
ENSDART00000104708
|
abhd12
|
abhydrolase domain containing 12 |
| chr18_+_14595546 | 0.18 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr11_+_5817202 | 0.16 |
ENSDART00000126084
|
ctdspl3
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 3 |
| chr10_-_22106331 | 0.16 |
ENSDART00000187654
|
BX005027.2
|
|
| chr13_+_49175624 | 0.16 |
ENSDART00000193550
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr3_+_57820913 | 0.15 |
ENSDART00000168101
|
CU571328.1
|
|
| chr12_+_46483618 | 0.15 |
ENSDART00000186970
|
BX005305.5
|
|
| chr21_-_588858 | 0.14 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr14_+_34951202 | 0.14 |
ENSDART00000047524
ENSDART00000115105 |
il12ba
|
interleukin 12Ba |
| chr5_+_71924175 | 0.14 |
ENSDART00000115182
ENSDART00000170215 |
nup214
|
nucleoporin 214 |
| chr12_+_46462090 | 0.13 |
ENSDART00000130748
|
BX005305.2
|
|
| chr17_+_31739418 | 0.13 |
ENSDART00000155073
ENSDART00000156180 |
arhgap5
|
Rho GTPase activating protein 5 |
| chr6_+_8630355 | 0.12 |
ENSDART00000161749
ENSDART00000193976 |
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr15_-_37436082 | 0.11 |
ENSDART00000192423
|
si:ch211-113j13.2
|
si:ch211-113j13.2 |
| chr17_-_5769196 | 0.11 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr4_+_26053044 | 0.11 |
ENSDART00000039877
|
SCYL2
|
si:ch211-244b2.1 |
| chr4_-_45028479 | 0.10 |
ENSDART00000143150
ENSDART00000076870 |
si:dkey-51d8.1
|
si:dkey-51d8.1 |
| chr1_+_49814461 | 0.09 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr11_+_36683859 | 0.09 |
ENSDART00000170102
|
si:ch211-11c3.12
|
si:ch211-11c3.12 |
| chr12_+_4686145 | 0.08 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr10_-_40617534 | 0.08 |
ENSDART00000148070
ENSDART00000131599 |
taar16g
|
trace amine associated receptor 16g |
| chr20_-_874807 | 0.07 |
ENSDART00000020506
|
snx14
|
sorting nexin 14 |
| chr16_-_12236362 | 0.07 |
ENSDART00000114759
|
lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
| chr13_+_49175947 | 0.07 |
ENSDART00000056927
|
egln1a
|
egl-9 family hypoxia-inducible factor 1a |
| chr4_+_19127973 | 0.07 |
ENSDART00000136611
|
si:dkey-21o22.2
|
si:dkey-21o22.2 |
| chr18_+_14595805 | 0.04 |
ENSDART00000167825
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr25_-_13549577 | 0.03 |
ENSDART00000166772
|
ano10b
|
anoctamin 10b |
| chr15_+_15856178 | 0.03 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr20_-_46249185 | 0.03 |
ENSDART00000100544
|
taar10
|
trace amine-associated receptor 10 |
| chr21_+_40302747 | 0.02 |
ENSDART00000174166
|
si:ch211-218m3.18
|
si:ch211-218m3.18 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmga1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 3.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.3 | 2.4 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.2 | 1.0 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.2 | 1.6 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.2 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.9 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 0.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.8 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.7 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.4 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.6 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.8 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.6 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.6 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.8 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.5 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.8 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.9 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.2 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.8 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.6 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 1.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.6 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.8 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 0.4 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 2.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 1.6 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.4 | 3.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.3 | 2.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.2 | 2.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 1.5 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.4 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 2.0 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.3 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.6 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.7 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.0 | 1.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.2 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.7 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 1.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 2.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |