Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
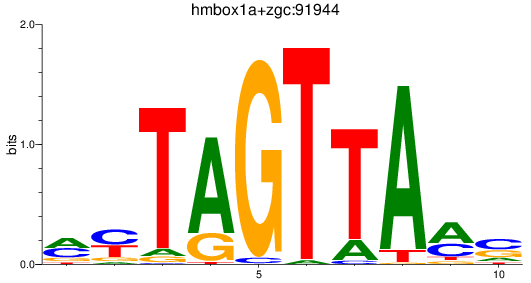
Results for hmbox1a+zgc:91944
Z-value: 0.73
Transcription factors associated with hmbox1a+zgc:91944
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hmbox1a
|
ENSDARG00000027082 | homeobox containing 1a |
|
zgc
|
ENSDARG00000035887 | 91944 |
|
hmbox1a
|
ENSDARG00000109287 | homeobox containing 1 b |
|
zgc
|
ENSDARG00000114642 | 91944 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CABZ01078261.1 | dr11_v1_chr20_+_49787584_49787584 | 0.47 | 4.3e-02 | Click! |
| zgc:91944 | dr11_v1_chr19_-_32600638_32600638 | -0.13 | 5.8e-01 | Click! |
| hmbox1a | dr11_v1_chr17_-_16324565_16324565 | 0.04 | 8.6e-01 | Click! |
Activity profile of hmbox1a+zgc:91944 motif
Sorted Z-values of hmbox1a+zgc:91944 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_71758307 | 2.19 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr11_+_43043171 | 1.52 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr7_+_35075847 | 1.44 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr4_-_77506362 | 1.31 |
ENSDART00000174387
ENSDART00000181181 |
CABZ01087415.1
|
|
| chr15_-_17074393 | 1.00 |
ENSDART00000155526
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr4_+_72798545 | 0.91 |
ENSDART00000181727
|
MYRFL
|
myelin regulatory factor-like |
| chr19_-_40192249 | 0.90 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr12_+_34896956 | 0.89 |
ENSDART00000055415
|
prph2a
|
peripherin 2a (retinal degeneration, slow) |
| chr3_+_39540014 | 0.86 |
ENSDART00000074848
|
zgc:165423
|
zgc:165423 |
| chr23_-_44786844 | 0.79 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr3_-_31079186 | 0.79 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr17_-_24919297 | 0.78 |
ENSDART00000154424
|
zgc:193593
|
zgc:193593 |
| chr17_-_36896560 | 0.71 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr8_+_39724138 | 0.70 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr24_+_9178064 | 0.69 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr7_+_2849020 | 0.68 |
ENSDART00000168695
|
BX004857.1
|
|
| chr23_-_11130683 | 0.68 |
ENSDART00000181189
|
cntn3a.2
|
contactin 3a, tandem duplicate 2 |
| chr19_+_9174166 | 0.67 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr4_+_76509294 | 0.67 |
ENSDART00000099899
|
ms4a17a.17
|
membrane-spanning 4-domains, subfamily A, member 17A.17 |
| chr3_-_18675688 | 0.66 |
ENSDART00000048218
|
slc5a11
|
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr25_+_35706493 | 0.64 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr20_-_47051996 | 0.62 |
ENSDART00000153330
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr16_+_45930962 | 0.59 |
ENSDART00000124689
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr25_+_25737386 | 0.57 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr1_-_9641845 | 0.57 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr5_-_12687357 | 0.56 |
ENSDART00000193206
|
aifm3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr3_+_49043917 | 0.56 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr8_-_37328112 | 0.55 |
ENSDART00000140365
|
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr17_-_15188440 | 0.54 |
ENSDART00000151885
|
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
| chr5_-_61624693 | 0.54 |
ENSDART00000141323
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr18_+_9615147 | 0.53 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr20_-_7133546 | 0.53 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr9_-_54248182 | 0.52 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr7_+_41295974 | 0.52 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr20_-_1378514 | 0.51 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr23_-_14766902 | 0.51 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr13_+_46718518 | 0.51 |
ENSDART00000160401
ENSDART00000182884 |
tmem63ba
|
transmembrane protein 63Ba |
| chr24_-_8887825 | 0.51 |
ENSDART00000066781
|
elovl2
|
ELOVL fatty acid elongase 2 |
| chr1_+_53374454 | 0.51 |
ENSDART00000038807
|
ucp1
|
uncoupling protein 1 |
| chr10_+_4874542 | 0.51 |
ENSDART00000101431
|
palm2
|
paralemmin 2 |
| chr21_-_22474362 | 0.50 |
ENSDART00000169659
|
myo5b
|
myosin VB |
| chr10_+_439692 | 0.49 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr18_+_11506561 | 0.49 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr18_-_341489 | 0.49 |
ENSDART00000168333
|
si:ch1073-83b15.1
|
si:ch1073-83b15.1 |
| chr21_-_42097736 | 0.49 |
ENSDART00000100000
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr7_-_71758613 | 0.48 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr13_+_1542493 | 0.47 |
ENSDART00000181968
|
CABZ01044281.1
|
|
| chr24_-_29868151 | 0.47 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr18_+_13077800 | 0.46 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr2_+_2503396 | 0.45 |
ENSDART00000168418
|
crhr2
|
corticotropin releasing hormone receptor 2 |
| chr18_-_2727764 | 0.45 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr9_-_34986827 | 0.44 |
ENSDART00000137862
|
si:ch211-160b11.4
|
si:ch211-160b11.4 |
| chr19_+_24394560 | 0.43 |
ENSDART00000142506
|
si:dkey-81h8.1
|
si:dkey-81h8.1 |
| chr5_+_20257225 | 0.43 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr7_+_19016023 | 0.43 |
ENSDART00000185212
|
CU972454.5
|
|
| chr19_-_25271155 | 0.43 |
ENSDART00000104027
|
rims3
|
regulating synaptic membrane exocytosis 3 |
| chr17_+_33375469 | 0.42 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr19_+_43669122 | 0.42 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr13_-_21660203 | 0.41 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr16_-_46587938 | 0.41 |
ENSDART00000181433
|
BX323793.1
|
|
| chr19_+_823945 | 0.41 |
ENSDART00000142287
|
ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr21_+_33459524 | 0.40 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr11_-_29650930 | 0.39 |
ENSDART00000166969
|
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr17_-_47142249 | 0.39 |
ENSDART00000184705
|
CU915775.1
|
|
| chr2_+_55914699 | 0.38 |
ENSDART00000157905
|
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr19_+_9295244 | 0.37 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr1_+_44395976 | 0.37 |
ENSDART00000159686
ENSDART00000189905 ENSDART00000025145 |
unc93b1
|
unc-93 homolog B1, TLR signaling regulator |
| chr14_-_29858883 | 0.36 |
ENSDART00000141034
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr13_-_37620091 | 0.36 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr11_+_11030908 | 0.35 |
ENSDART00000111091
|
pla2r1
|
phospholipase A2 receptor 1 |
| chr10_-_44508249 | 0.34 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr23_-_30960506 | 0.33 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr21_+_5635420 | 0.33 |
ENSDART00000168158
|
shroom3
|
shroom family member 3 |
| chr2_-_48966431 | 0.32 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr2_-_3403020 | 0.32 |
ENSDART00000092741
|
snap47
|
synaptosomal-associated protein, 47 |
| chr12_-_13650344 | 0.32 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr14_+_21222287 | 0.32 |
ENSDART00000159905
|
si:ch211-175m2.4
|
si:ch211-175m2.4 |
| chr24_-_7321928 | 0.32 |
ENSDART00000167570
ENSDART00000045150 |
actr3b
|
ARP3 actin related protein 3 homolog B |
| chr19_+_31585917 | 0.32 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr12_+_29054907 | 0.32 |
ENSDART00000152936
|
gabrz
|
gamma-aminobutyric acid (GABA) A receptor, zeta |
| chr11_+_1764203 | 0.30 |
ENSDART00000173023
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr12_+_30200652 | 0.30 |
ENSDART00000193405
ENSDART00000132335 |
ablim1b
|
actin binding LIM protein 1b |
| chr14_+_23709134 | 0.30 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr7_+_13609457 | 0.30 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr23_+_26946429 | 0.30 |
ENSDART00000185564
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr8_+_53064920 | 0.30 |
ENSDART00000164823
|
nadka
|
NAD kinase a |
| chr10_-_28513861 | 0.29 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr7_+_36467796 | 0.29 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr19_+_31585341 | 0.29 |
ENSDART00000052185
|
gmnn
|
geminin, DNA replication inhibitor |
| chr11_-_1400507 | 0.28 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr22_-_8388678 | 0.28 |
ENSDART00000184389
|
CABZ01061498.1
|
|
| chr12_+_10631266 | 0.28 |
ENSDART00000161455
|
csf3a
|
colony stimulating factor 3 (granulocyte) a |
| chr18_-_502722 | 0.28 |
ENSDART00000185757
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr7_+_65261576 | 0.28 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr12_-_19153177 | 0.28 |
ENSDART00000057125
|
tefa
|
thyrotrophic embryonic factor a |
| chr13_-_36050303 | 0.27 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr12_-_20665164 | 0.27 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr12_-_26538823 | 0.27 |
ENSDART00000143213
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr8_+_10339869 | 0.27 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr16_-_48071632 | 0.27 |
ENSDART00000148564
|
CSMD3 (1 of many)
|
si:ch211-236p22.1 |
| chr12_+_35654749 | 0.26 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr4_+_26056548 | 0.26 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr7_-_38570878 | 0.26 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr6_+_10450000 | 0.26 |
ENSDART00000151288
ENSDART00000187431 ENSDART00000192474 ENSDART00000188214 ENSDART00000184766 ENSDART00000190082 |
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr13_+_8693410 | 0.25 |
ENSDART00000138448
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr14_+_16151636 | 0.25 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr24_-_18809433 | 0.25 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr3_-_11878490 | 0.25 |
ENSDART00000129961
|
coro7
|
coronin 7 |
| chr7_-_18416741 | 0.24 |
ENSDART00000097882
|
sstr1b
|
somatostatin receptor 1b |
| chr23_-_23160088 | 0.24 |
ENSDART00000141853
|
noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr24_-_20321012 | 0.24 |
ENSDART00000163683
|
map3k20
|
mitogen-activated protein kinase kinase kinase 20 |
| chr3_+_15805917 | 0.24 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr2_+_49713592 | 0.24 |
ENSDART00000189624
|
BX323861.3
|
|
| chr13_-_8279533 | 0.24 |
ENSDART00000109666
|
si:ch211-250c4.5
|
si:ch211-250c4.5 |
| chr14_-_29859067 | 0.24 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr17_+_25197180 | 0.24 |
ENSDART00000148775
|
arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr8_+_14890821 | 0.24 |
ENSDART00000190966
|
soat1
|
sterol O-acyltransferase 1 |
| chr24_-_35269991 | 0.24 |
ENSDART00000185424
|
sntg1
|
syntrophin, gamma 1 |
| chr6_-_30658755 | 0.23 |
ENSDART00000065215
ENSDART00000181302 |
lurap1
|
leucine rich adaptor protein 1 |
| chr8_+_7144066 | 0.22 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr4_-_45100253 | 0.22 |
ENSDART00000163870
|
si:dkey-51d8.3
|
si:dkey-51d8.3 |
| chr2_-_43191465 | 0.22 |
ENSDART00000025254
|
crema
|
cAMP responsive element modulator a |
| chr6_-_30683637 | 0.21 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr8_+_30699429 | 0.21 |
ENSDART00000005345
|
upb1
|
ureidopropionase, beta |
| chr20_-_30377221 | 0.21 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr1_-_11372456 | 0.21 |
ENSDART00000144164
ENSDART00000141238 |
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr3_-_30186296 | 0.21 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr7_+_53199763 | 0.20 |
ENSDART00000160097
|
cdh28
|
cadherin 28 |
| chr8_-_11546175 | 0.20 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr2_+_23677179 | 0.20 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr10_-_22912255 | 0.20 |
ENSDART00000131992
|
si:ch1073-143l10.2
|
si:ch1073-143l10.2 |
| chr11_-_40030139 | 0.20 |
ENSDART00000021916
|
uts2b
|
urotensin 2, beta |
| chr9_+_29643036 | 0.20 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr5_+_19448078 | 0.20 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr10_+_35952532 | 0.20 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr13_+_40019001 | 0.19 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr12_+_40945319 | 0.19 |
ENSDART00000183435
|
CDH18
|
cadherin 18 |
| chr7_-_27037990 | 0.19 |
ENSDART00000173561
|
nucb2a
|
nucleobindin 2a |
| chr2_-_45331115 | 0.19 |
ENSDART00000158003
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr21_-_4923427 | 0.19 |
ENSDART00000181489
|
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr18_-_30021479 | 0.19 |
ENSDART00000144562
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr2_+_29491314 | 0.19 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr23_+_26946744 | 0.19 |
ENSDART00000115141
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr25_-_19486399 | 0.19 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr9_-_49805218 | 0.18 |
ENSDART00000179290
|
GALNT3
|
polypeptide N-acetylgalactosaminyltransferase 3 |
| chr8_+_21384288 | 0.18 |
ENSDART00000079263
|
fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_-_36230146 | 0.18 |
ENSDART00000135888
ENSDART00000189782 |
rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr21_-_37973081 | 0.17 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr19_-_22328154 | 0.17 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr2_-_30460293 | 0.17 |
ENSDART00000113193
|
cbln2a
|
cerebellin 2a precursor |
| chr20_-_456998 | 0.17 |
ENSDART00000149973
|
frk
|
fyn-related Src family tyrosine kinase |
| chr2_+_11029138 | 0.16 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr6_-_48473694 | 0.16 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr20_+_53522059 | 0.16 |
ENSDART00000147570
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr5_-_13564961 | 0.16 |
ENSDART00000146827
|
si:ch211-230g14.3
|
si:ch211-230g14.3 |
| chr4_+_20566371 | 0.16 |
ENSDART00000127576
|
BX248410.1
|
|
| chr13_+_48359573 | 0.16 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr18_-_8857137 | 0.16 |
ENSDART00000126331
|
prrt4
|
proline-rich transmembrane protein 4 |
| chr17_-_25382367 | 0.15 |
ENSDART00000162306
ENSDART00000165282 |
lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr12_+_35011899 | 0.15 |
ENSDART00000153007
ENSDART00000153020 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr23_+_41679586 | 0.15 |
ENSDART00000067662
|
CU914487.1
|
|
| chr2_-_24061575 | 0.15 |
ENSDART00000089234
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr4_+_71989418 | 0.15 |
ENSDART00000170996
|
parp11
|
poly(ADP-ribose) polymerase family member 11 |
| chr10_-_22095505 | 0.15 |
ENSDART00000140210
|
ponzr10
|
plac8 onzin related protein 10 |
| chr22_-_16755885 | 0.15 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr3_-_60827402 | 0.15 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr13_+_15190677 | 0.15 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr15_+_44366556 | 0.14 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr7_-_16195144 | 0.14 |
ENSDART00000173492
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr6_-_31325400 | 0.14 |
ENSDART00000188869
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr11_+_44622019 | 0.14 |
ENSDART00000159618
|
rbm34
|
RNA binding motif protein 34 |
| chr16_-_43011470 | 0.14 |
ENSDART00000131898
ENSDART00000142003 ENSDART00000017966 |
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr13_-_18548729 | 0.14 |
ENSDART00000187686
|
BX908770.1
|
|
| chr6_+_4160579 | 0.14 |
ENSDART00000105278
ENSDART00000187932 ENSDART00000111817 |
trim25l
|
tripartite motif containing 25, like |
| chr22_+_19366866 | 0.14 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr21_-_4922981 | 0.13 |
ENSDART00000191819
ENSDART00000113556 ENSDART00000193269 ENSDART00000182961 |
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr14_+_31498439 | 0.13 |
ENSDART00000173281
|
phf6
|
PHD finger protein 6 |
| chr13_-_31938512 | 0.13 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr8_-_36112278 | 0.13 |
ENSDART00000183283
|
CT583723.1
|
|
| chr18_-_31051847 | 0.13 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr24_+_12133814 | 0.13 |
ENSDART00000158562
ENSDART00000159029 ENSDART00000168248 |
lztfl1
|
leucine zipper transcription factor-like 1 |
| chr4_-_1908179 | 0.13 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr22_-_17586064 | 0.13 |
ENSDART00000060786
ENSDART00000188303 ENSDART00000181212 ENSDART00000181951 |
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr2_-_2957970 | 0.13 |
ENSDART00000162505
|
si:ch1073-82l19.1
|
si:ch1073-82l19.1 |
| chr10_+_45302425 | 0.13 |
ENSDART00000159954
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr25_+_4750972 | 0.13 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr22_-_17585618 | 0.13 |
ENSDART00000183123
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr5_-_67799821 | 0.12 |
ENSDART00000017881
|
eif4e1b
|
eukaryotic translation initiation factor 4E family member 1B |
| chr17_-_37184655 | 0.12 |
ENSDART00000180447
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr5_+_36439405 | 0.12 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr1_-_513762 | 0.12 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr15_-_23814330 | 0.12 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr16_+_43401005 | 0.12 |
ENSDART00000110994
|
sqlea
|
squalene epoxidase a |
| chr4_-_14486822 | 0.12 |
ENSDART00000048821
|
plxnb2a
|
plexin b2a |
| chr17_+_17861681 | 0.12 |
ENSDART00000123311
|
ism2a
|
isthmin 2a |
| chr7_+_65813301 | 0.12 |
ENSDART00000187449
ENSDART00000179834 ENSDART00000082740 ENSDART00000178627 |
tead1b
|
TEA domain family member 1b |
| chr6_+_37441789 | 0.12 |
ENSDART00000157317
|
gtpbp6
|
GTP binding protein 6 (putative) |
| chr17_+_5985933 | 0.12 |
ENSDART00000190844
|
zgc:194275
|
zgc:194275 |
| chr19_-_5669122 | 0.12 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr5_-_11809404 | 0.11 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hmbox1a+zgc:91944
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.5 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 3 signaling pathway(GO:0034138) toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.3 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.3 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 0.3 | GO:0061113 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.2 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.9 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.4 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0019483 | beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) |
| 0.0 | 0.2 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.4 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.5 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.6 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.2 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 1.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 0.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 2.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.1 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.0 | 0.3 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.5 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 2.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.5 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.3 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.3 | GO:0031956 | medium-chain fatty acid-CoA ligase activity(GO:0031956) butyrate-CoA ligase activity(GO:0047760) |
| 0.1 | 0.2 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.3 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.1 | 0.2 | GO:0032404 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 2.6 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |