Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
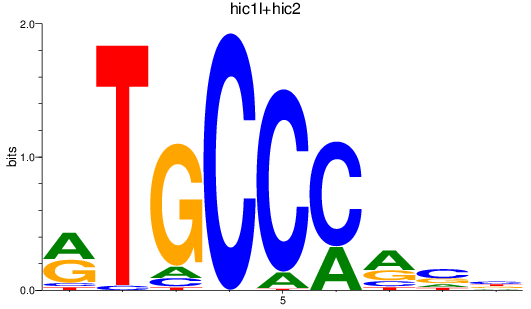
Results for hic1l+hic2
Z-value: 0.59
Transcription factors associated with hic1l+hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hic1l
|
ENSDARG00000045660 | hypermethylated in cancer 1 like |
|
hic2
|
ENSDARG00000100497 | hypermethylated in cancer 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hic1l | dr11_v1_chr25_-_25575241_25575241 | -0.74 | 3.2e-04 | Click! |
| hic2 | dr11_v1_chr10_+_3145707_3145707 | -0.71 | 7.3e-04 | Click! |
Activity profile of hic1l+hic2 motif
Sorted Z-values of hic1l+hic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_6159162 | 1.49 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr1_-_55810730 | 1.27 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr5_+_32815745 | 1.03 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr25_+_31222069 | 1.02 |
ENSDART00000159373
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr12_+_28367557 | 1.02 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr18_-_16123222 | 1.02 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr17_-_19344999 | 1.01 |
ENSDART00000138315
|
gsc
|
goosecoid |
| chr5_-_57641257 | 0.99 |
ENSDART00000149282
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr5_+_15992655 | 0.92 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr21_+_5169154 | 0.88 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr7_+_29952169 | 0.81 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr3_-_24980067 | 0.77 |
ENSDART00000048871
|
desi1a
|
desumoylating isopeptidase 1a |
| chr23_+_2669 | 0.77 |
ENSDART00000011146
|
twist3
|
twist3 |
| chr3_+_23221047 | 0.77 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr8_+_38417461 | 0.76 |
ENSDART00000132718
|
nkx6.3
|
NK6 homeobox 3 |
| chr6_-_49634787 | 0.76 |
ENSDART00000188538
|
BX323459.1
|
|
| chr25_-_7686201 | 0.76 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr12_-_22524388 | 0.74 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr20_+_4392687 | 0.72 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr21_+_45733871 | 0.72 |
ENSDART00000187285
ENSDART00000193018 |
zgc:77058
|
zgc:77058 |
| chr12_-_20362041 | 0.71 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr17_+_23937262 | 0.71 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr4_-_8040436 | 0.70 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr2_-_44720551 | 0.66 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr8_-_33114202 | 0.66 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr3_-_32170850 | 0.66 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr3_+_34821327 | 0.66 |
ENSDART00000055262
|
cdk5r1a
|
cyclin-dependent kinase 5, regulatory subunit 1a (p35) |
| chr12_+_42574148 | 0.65 |
ENSDART00000157855
|
ebf3a
|
early B cell factor 3a |
| chr7_+_29952719 | 0.65 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr25_+_7229046 | 0.62 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr1_-_38538284 | 0.62 |
ENSDART00000009777
|
glra3
|
glycine receptor, alpha 3 |
| chr23_-_21797517 | 0.61 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr16_+_35344031 | 0.59 |
ENSDART00000167140
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr2_+_11206317 | 0.58 |
ENSDART00000144982
|
lhx8a
|
LIM homeobox 8a |
| chr16_+_31853919 | 0.54 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr1_-_23040358 | 0.54 |
ENSDART00000146871
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr8_-_38616712 | 0.53 |
ENSDART00000141827
|
si:ch211-198d23.1
|
si:ch211-198d23.1 |
| chr17_-_12926456 | 0.51 |
ENSDART00000044126
|
INSM2
|
INSM transcriptional repressor 2 |
| chr19_-_41052240 | 0.50 |
ENSDART00000131865
|
sgce
|
sarcoglycan, epsilon |
| chr19_-_26235219 | 0.50 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr4_-_14915268 | 0.49 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr23_-_27875140 | 0.48 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr25_-_27541621 | 0.46 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr3_-_61592417 | 0.45 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr10_+_43188678 | 0.44 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr24_-_29822913 | 0.44 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr8_-_14144707 | 0.43 |
ENSDART00000148061
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr5_-_26181863 | 0.43 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr25_-_13659249 | 0.42 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr5_-_38122126 | 0.42 |
ENSDART00000141791
ENSDART00000170528 |
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr21_+_25375172 | 0.41 |
ENSDART00000078748
|
or132-5
|
odorant receptor, family H, subfamily 132, member 5 |
| chr10_-_32920690 | 0.41 |
ENSDART00000136245
|
cux1a
|
cut-like homeobox 1a |
| chr17_+_32360673 | 0.40 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr16_+_43347966 | 0.40 |
ENSDART00000171308
|
zmp:0000000930
|
zmp:0000000930 |
| chr22_+_7497319 | 0.38 |
ENSDART00000034564
|
CELA1 (1 of many)
|
zgc:92511 |
| chr19_-_41052408 | 0.37 |
ENSDART00000151120
|
sgce
|
sarcoglycan, epsilon |
| chr22_+_7742211 | 0.37 |
ENSDART00000140896
|
CELA1 (1 of many)
|
zgc:92511 |
| chr16_+_12836143 | 0.37 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr9_+_29643036 | 0.37 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr12_+_31729075 | 0.37 |
ENSDART00000152973
|
RNF157
|
si:dkey-49c17.3 |
| chr17_+_26657896 | 0.36 |
ENSDART00000152037
|
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr21_+_18877130 | 0.36 |
ENSDART00000136893
|
si:dkey-65l23.2
|
si:dkey-65l23.2 |
| chr13_-_37383625 | 0.36 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr2_+_31475772 | 0.36 |
ENSDART00000130722
|
cacnb2b
|
calcium channel, voltage-dependent, beta 2b |
| chr5_-_38121612 | 0.34 |
ENSDART00000159543
|
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr1_+_44992207 | 0.34 |
ENSDART00000172357
|
tmem132a
|
transmembrane protein 132A |
| chr10_-_34867401 | 0.33 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr5_-_30984271 | 0.33 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr25_-_19068557 | 0.32 |
ENSDART00000184780
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr13_+_7292061 | 0.32 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr20_+_40766645 | 0.32 |
ENSDART00000144401
|
tbc1d32
|
TBC1 domain family, member 32 |
| chr23_-_1587955 | 0.32 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr22_-_7129631 | 0.32 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr14_-_9199968 | 0.32 |
ENSDART00000146113
|
arhgef9b
|
Cdc42 guanine nucleotide exchange factor (GEF) 9b |
| chr19_+_47311869 | 0.31 |
ENSDART00000136647
|
ext1c
|
exostoses (multiple) 1c |
| chr5_-_29587351 | 0.31 |
ENSDART00000136446
ENSDART00000051434 |
entpd2a.1
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 1 |
| chr17_+_52823015 | 0.31 |
ENSDART00000160507
ENSDART00000186979 |
meis2a
|
Meis homeobox 2a |
| chr12_-_47580562 | 0.30 |
ENSDART00000153431
|
rgs7b
|
regulator of G protein signaling 7b |
| chr16_-_52916248 | 0.30 |
ENSDART00000113931
|
zdhhc11
|
zinc finger, DHHC-type containing 11 |
| chr18_-_50845804 | 0.30 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr25_-_3087556 | 0.29 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
| chr13_+_32148338 | 0.29 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr9_-_33107237 | 0.29 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr16_-_37964325 | 0.29 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr5_-_69041102 | 0.29 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr12_-_44151296 | 0.28 |
ENSDART00000168734
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr21_-_40562705 | 0.28 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr10_+_34623183 | 0.27 |
ENSDART00000114630
|
nbeaa
|
neurobeachin a |
| chr23_-_6765653 | 0.27 |
ENSDART00000192310
|
FP102169.1
|
|
| chr16_+_35343493 | 0.27 |
ENSDART00000171269
|
si:dkey-34d22.1
|
si:dkey-34d22.1 |
| chr2_-_36918709 | 0.26 |
ENSDART00000084876
|
zgc:153654
|
zgc:153654 |
| chr24_-_39858710 | 0.26 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr2_+_26240339 | 0.26 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr10_-_8129175 | 0.26 |
ENSDART00000133921
|
ndufa9b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9b |
| chr7_+_26172396 | 0.26 |
ENSDART00000173975
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr23_+_27740788 | 0.25 |
ENSDART00000053871
|
dhh
|
desert hedgehog |
| chr23_-_32404022 | 0.25 |
ENSDART00000156387
ENSDART00000155508 |
si:ch211-66i15.4
|
si:ch211-66i15.4 |
| chr15_+_31471808 | 0.25 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr7_-_46782264 | 0.24 |
ENSDART00000166565
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr4_-_5597167 | 0.24 |
ENSDART00000132431
|
vegfab
|
vascular endothelial growth factor Ab |
| chr15_-_11956981 | 0.24 |
ENSDART00000164163
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr12_+_31729498 | 0.24 |
ENSDART00000188546
ENSDART00000182562 ENSDART00000186147 |
RNF157
|
si:dkey-49c17.3 |
| chr15_-_669476 | 0.23 |
ENSDART00000153687
ENSDART00000030603 |
si:ch211-210b2.2
|
si:ch211-210b2.2 |
| chr2_-_41942666 | 0.23 |
ENSDART00000075673
|
ebi3
|
Epstein-Barr virus induced 3 |
| chr17_+_15845765 | 0.23 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr6_-_59381391 | 0.23 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr14_-_14746767 | 0.23 |
ENSDART00000183755
ENSDART00000190938 |
ogt.1
|
O-linked N-acetylglucosamine (GlcNAc) transferase, tandem duplicate 1 |
| chr6_+_29402997 | 0.22 |
ENSDART00000104298
|
ndufb5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 5 |
| chr5_-_6508250 | 0.22 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr17_+_27134806 | 0.22 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr5_-_67145505 | 0.22 |
ENSDART00000011295
|
rom1a
|
retinal outer segment membrane protein 1a |
| chr2_-_37889111 | 0.21 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr11_+_4953964 | 0.21 |
ENSDART00000092552
|
ptprga
|
protein tyrosine phosphatase, receptor type, g a |
| chr7_-_22132265 | 0.20 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr5_+_39537195 | 0.20 |
ENSDART00000051256
|
fgf5
|
fibroblast growth factor 5 |
| chr5_-_57820873 | 0.19 |
ENSDART00000089961
|
sik2a
|
salt-inducible kinase 2a |
| chr1_+_10720294 | 0.19 |
ENSDART00000139387
|
atp1b1b
|
ATPase Na+/K+ transporting subunit beta 1b |
| chr4_-_8030583 | 0.18 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr7_-_43857938 | 0.18 |
ENSDART00000183602
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr14_-_897874 | 0.18 |
ENSDART00000167395
|
rgs14a
|
regulator of G protein signaling 14a |
| chr16_-_53800047 | 0.18 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr9_-_40935934 | 0.18 |
ENSDART00000155604
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr12_+_28995942 | 0.18 |
ENSDART00000076334
|
valopb
|
vertebrate ancient long opsin b |
| chr10_+_7182423 | 0.18 |
ENSDART00000186788
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr14_+_50807558 | 0.18 |
ENSDART00000183649
|
si:ch211-59p23.1
|
si:ch211-59p23.1 |
| chr14_-_5677979 | 0.16 |
ENSDART00000182156
|
tlx2
|
T cell leukemia homeobox 2 |
| chr18_-_12612699 | 0.16 |
ENSDART00000090335
|
hipk2
|
homeodomain interacting protein kinase 2 |
| chr20_+_23185530 | 0.16 |
ENSDART00000142346
|
lrrc66
|
leucine rich repeat containing 66 |
| chr20_+_27003713 | 0.15 |
ENSDART00000153232
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr24_-_33291784 | 0.15 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr3_+_13830599 | 0.15 |
ENSDART00000159282
|
ptger1b
|
prostaglandin E receptor 1b (subtype EP1) |
| chr2_-_43605556 | 0.15 |
ENSDART00000084223
|
epc1b
|
enhancer of polycomb homolog 1 (Drosophila) b |
| chr13_-_45811137 | 0.15 |
ENSDART00000190512
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr12_-_44010532 | 0.15 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr25_-_16146851 | 0.15 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr13_-_9061944 | 0.15 |
ENSDART00000164186
ENSDART00000102051 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr15_+_42626957 | 0.15 |
ENSDART00000154754
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr6_+_13726844 | 0.15 |
ENSDART00000055833
|
wnt6a
|
wingless-type MMTV integration site family, member 6a |
| chr10_-_26179805 | 0.15 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr17_+_14886828 | 0.14 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr4_-_25485404 | 0.14 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr5_+_57641554 | 0.14 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr19_+_14109348 | 0.14 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr7_-_65191937 | 0.13 |
ENSDART00000173234
|
pkd1l2a
|
polycystic kidney disease 1 like 2a |
| chr19_+_4463389 | 0.13 |
ENSDART00000168805
|
kcnk9
|
potassium channel, subfamily K, member 9 |
| chr10_+_41199660 | 0.13 |
ENSDART00000125314
|
adrb3b
|
adrenoceptor beta 3b |
| chr19_-_24958393 | 0.13 |
ENSDART00000098592
|
si:ch211-195b13.6
|
si:ch211-195b13.6 |
| chr18_+_20677090 | 0.13 |
ENSDART00000060243
|
cftr
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr15_-_13254480 | 0.12 |
ENSDART00000190499
|
zgc:172282
|
zgc:172282 |
| chr5_-_26834511 | 0.12 |
ENSDART00000136713
ENSDART00000192932 ENSDART00000113246 |
si:ch211-102c2.4
|
si:ch211-102c2.4 |
| chr9_+_29994439 | 0.12 |
ENSDART00000012447
|
tmem30c
|
transmembrane protein 30C |
| chr2_+_24936766 | 0.12 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr25_+_30238021 | 0.11 |
ENSDART00000123220
|
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr8_+_29267093 | 0.10 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr17_+_53294228 | 0.10 |
ENSDART00000158172
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr16_-_25663846 | 0.09 |
ENSDART00000031304
|
derl1
|
derlin 1 |
| chr2_+_30547018 | 0.09 |
ENSDART00000193747
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr14_-_448182 | 0.08 |
ENSDART00000180018
|
FAT4
|
FAT atypical cadherin 4 |
| chr21_+_13244450 | 0.08 |
ENSDART00000146062
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr8_+_35664152 | 0.08 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr20_+_54280549 | 0.08 |
ENSDART00000151048
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr7_+_21887307 | 0.07 |
ENSDART00000052871
|
pop7
|
POP7 homolog, ribonuclease P/MRP subunit |
| chr8_-_7475917 | 0.07 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr10_-_22828302 | 0.07 |
ENSDART00000079459
ENSDART00000100468 |
per1a
|
period circadian clock 1a |
| chr7_+_34786591 | 0.07 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr15_-_37779978 | 0.07 |
ENSDART00000157202
|
si:dkey-42l23.3
|
si:dkey-42l23.3 |
| chr19_+_19241372 | 0.07 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr9_-_34885902 | 0.07 |
ENSDART00000137504
|
p2ry8
|
P2Y receptor family member 8 |
| chr3_-_61065055 | 0.06 |
ENSDART00000074199
|
cacng1b
|
calcium channel, voltage-dependent, gamma subunit 1b |
| chr7_+_35268880 | 0.06 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr15_+_33851280 | 0.06 |
ENSDART00000162308
|
inpp5ka
|
inositol polyphosphate-5-phosphatase Ka |
| chr9_-_19610436 | 0.06 |
ENSDART00000108697
|
fgf9
|
fibroblast growth factor 9 |
| chr22_+_19262161 | 0.05 |
ENSDART00000163654
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr9_+_38737924 | 0.05 |
ENSDART00000147652
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr15_-_38202630 | 0.05 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr4_+_74396786 | 0.05 |
ENSDART00000127501
ENSDART00000174347 |
zmp:0000001020
|
zmp:0000001020 |
| chr10_+_42691210 | 0.05 |
ENSDART00000193813
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr12_+_22680115 | 0.04 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr15_-_17138640 | 0.04 |
ENSDART00000080777
|
mrpl28
|
mitochondrial ribosomal protein L28 |
| chr2_+_54827391 | 0.04 |
ENSDART00000167269
|
twsg1a
|
twisted gastrulation BMP signaling modulator 1a |
| chr2_+_30531726 | 0.04 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr22_-_9618867 | 0.04 |
ENSDART00000131246
|
si:ch73-286h23.4
|
si:ch73-286h23.4 |
| chr15_-_28082310 | 0.04 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr16_-_41807397 | 0.04 |
ENSDART00000138577
|
si:dkey-199f5.7
|
si:dkey-199f5.7 |
| chr14_-_41535822 | 0.04 |
ENSDART00000149407
|
itga6l
|
integrin, alpha 6, like |
| chr12_+_24952902 | 0.03 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr12_-_4243268 | 0.03 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr18_+_33100606 | 0.03 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr15_+_25158104 | 0.03 |
ENSDART00000128267
|
slc35f2l
|
info solute carrier family 35, member F2, like |
| chr23_-_1144425 | 0.02 |
ENSDART00000098271
|
psmb11b
|
proteasome subunit beta11b |
| chr22_+_5123479 | 0.02 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr7_+_17938128 | 0.02 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr19_-_13962469 | 0.02 |
ENSDART00000159062
|
paqr7a
|
progestin and adipoQ receptor family member VII, a |
| chr14_+_25464681 | 0.01 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr21_-_26520629 | 0.01 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr22_+_29009541 | 0.01 |
ENSDART00000169449
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr21_+_21612214 | 0.01 |
ENSDART00000008099
|
b9d2
|
B9 domain containing 2 |
| chr7_+_16963091 | 0.01 |
ENSDART00000173770
|
nav2a
|
neuron navigator 2a |
| chr12_-_1951233 | 0.01 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr2_-_23600821 | 0.00 |
ENSDART00000146217
|
BX677666.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of hic1l+hic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.2 | 0.9 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.2 | 1.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.8 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.2 | 1.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.5 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 0.4 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.3 | GO:1901006 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.2 | GO:0010359 | regulation of anion channel activity(GO:0010359) |
| 0.1 | 0.2 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.3 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.3 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.6 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.3 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.6 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.4 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.4 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 1.5 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.5 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.4 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 1.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.0 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.1 | 0.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.0 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |