Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
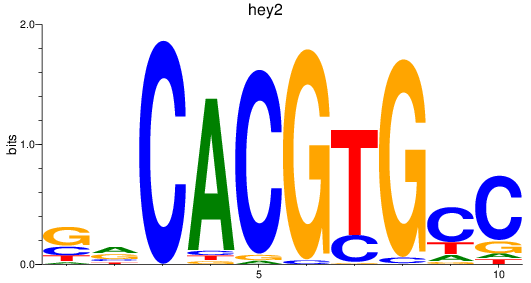
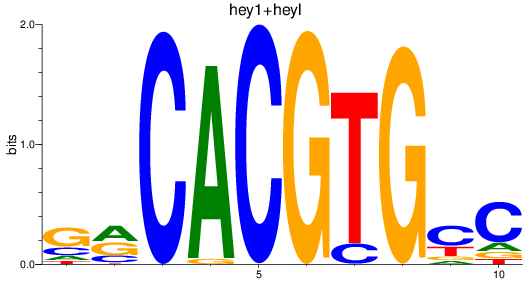
Results for hey2_hey1+heyl
Z-value: 1.14
Transcription factors associated with hey2_hey1+heyl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hey2
|
ENSDARG00000013441 | hes-related family bHLH transcription factor with YRPW motif 2 |
|
heyl
|
ENSDARG00000055798 | hes related family bHLH transcription factor with YRPW motif like |
|
hey1
|
ENSDARG00000070538 | hes-related family bHLH transcription factor with YRPW motif 1 |
|
heyl
|
ENSDARG00000112770 | hes related family bHLH transcription factor with YRPW motif like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hey2 | dr11_v1_chr20_-_39596338_39596338 | -0.88 | 8.8e-07 | Click! |
| hey1 | dr11_v1_chr19_-_31802296_31802296 | -0.54 | 1.7e-02 | Click! |
| heyl | dr11_v1_chr19_+_32553874_32553874 | -0.10 | 6.8e-01 | Click! |
Activity profile of hey2_hey1+heyl motif
Sorted Z-values of hey2_hey1+heyl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_13580348 | 8.33 |
ENSDART00000067160
|
opn1sw1
|
opsin 1 (cone pigments), short-wave-sensitive 1 |
| chr3_+_23488652 | 6.39 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr1_-_54765262 | 5.77 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr8_-_21372446 | 5.68 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr8_-_25247284 | 4.34 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr17_+_25414033 | 3.97 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr13_-_293250 | 3.94 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr14_+_35748385 | 3.92 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr2_+_21090317 | 3.90 |
ENSDART00000109568
ENSDART00000139633 |
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr14_+_35748206 | 3.63 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr3_+_1015867 | 3.42 |
ENSDART00000109912
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr11_-_3552067 | 3.35 |
ENSDART00000163656
|
CAMK2N1
|
si:dkey-33m11.6 |
| chr7_-_73752955 | 3.34 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr10_+_17026870 | 3.30 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr13_+_46803979 | 3.17 |
ENSDART00000159260
|
CU695232.1
|
|
| chr21_-_42100471 | 3.08 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr18_-_40684756 | 3.05 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr14_+_46313135 | 3.04 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr23_-_24856025 | 3.00 |
ENSDART00000142171
|
syt6a
|
synaptotagmin VIa |
| chr14_+_41409697 | 2.94 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr8_-_18239494 | 2.90 |
ENSDART00000079989
ENSDART00000022959 |
guk1b
|
guanylate kinase 1b |
| chr6_-_38818582 | 2.88 |
ENSDART00000149833
|
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr20_+_35473288 | 2.86 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr16_-_45069882 | 2.86 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr21_-_41588129 | 2.81 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr8_-_23081511 | 2.79 |
ENSDART00000142015
ENSDART00000135764 ENSDART00000147021 |
si:dkey-70p6.1
|
si:dkey-70p6.1 |
| chr7_+_38717624 | 2.77 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr10_+_158590 | 2.77 |
ENSDART00000081982
|
KCNJ15
|
potassium voltage-gated channel subfamily J member 15 |
| chr21_+_6290566 | 2.62 |
ENSDART00000161647
|
fnbp1b
|
formin binding protein 1b |
| chr6_-_60147517 | 2.54 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
| chr18_+_10784730 | 2.50 |
ENSDART00000028938
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr19_+_43563179 | 2.49 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr18_-_21218851 | 2.45 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr15_+_1148074 | 2.45 |
ENSDART00000152638
ENSDART00000152466 ENSDART00000188011 |
mlf1
|
myeloid leukemia factor 1 |
| chr20_+_50115335 | 2.42 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr1_-_50859053 | 2.41 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr7_-_823304 | 2.39 |
ENSDART00000173694
|
si:cabz01076231.1
|
si:cabz01076231.1 |
| chr17_-_37195163 | 2.24 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr24_+_39135419 | 2.22 |
ENSDART00000180941
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr10_-_41450367 | 2.20 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr9_-_22281854 | 2.19 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr23_+_40452157 | 2.13 |
ENSDART00000113106
ENSDART00000140136 |
soga3b
|
SOGA family member 3b |
| chr11_+_45299447 | 2.13 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr15_+_16897554 | 2.11 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr9_+_42066030 | 2.05 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr21_+_31150438 | 2.03 |
ENSDART00000065366
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_+_42469058 | 2.02 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr25_+_37443194 | 2.00 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr5_-_26950374 | 1.99 |
ENSDART00000050542
|
htra4
|
HtrA serine peptidase 4 |
| chr11_+_29770966 | 1.97 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr5_+_26075230 | 1.97 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr21_+_31150773 | 1.96 |
ENSDART00000126205
|
st6gal1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr19_-_8940068 | 1.94 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr17_-_37195354 | 1.93 |
ENSDART00000190963
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr9_-_22240052 | 1.92 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr5_-_69314495 | 1.90 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr25_+_3328487 | 1.90 |
ENSDART00000181143
|
ldhbb
|
lactate dehydrogenase Bb |
| chr20_-_27733683 | 1.89 |
ENSDART00000103317
ENSDART00000138139 |
zgc:153157
|
zgc:153157 |
| chr22_-_12160283 | 1.87 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr12_+_24342303 | 1.86 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr20_+_34915945 | 1.84 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr1_+_9004719 | 1.84 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr3_+_32526263 | 1.84 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr9_-_22232902 | 1.83 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr21_+_11969603 | 1.82 |
ENSDART00000142247
ENSDART00000140652 |
mlnl
|
motilin-like |
| chr4_+_5180650 | 1.81 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr17_+_43843536 | 1.80 |
ENSDART00000164097
|
FO704777.1
|
|
| chr19_+_22850657 | 1.79 |
ENSDART00000130472
|
pkhd1l1
|
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
| chr11_-_3959889 | 1.76 |
ENSDART00000159683
|
pbrm1
|
polybromo 1 |
| chr18_+_6479963 | 1.75 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr8_-_42238543 | 1.72 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr25_+_33192796 | 1.70 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr6_-_44711942 | 1.70 |
ENSDART00000055035
|
cntn3b
|
contactin 3b |
| chr14_+_2095394 | 1.64 |
ENSDART00000186847
|
CT573264.2
|
|
| chr3_+_32526799 | 1.63 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr21_+_6780340 | 1.63 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr3_+_1223824 | 1.63 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr5_-_18897482 | 1.61 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr2_+_20331445 | 1.59 |
ENSDART00000186880
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr25_+_37268900 | 1.59 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr6_+_22597362 | 1.59 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr15_-_44077937 | 1.58 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr25_-_21092222 | 1.56 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr21_+_6291027 | 1.53 |
ENSDART00000180467
ENSDART00000184952 ENSDART00000184006 |
fnbp1b
|
formin binding protein 1b |
| chr20_-_116999 | 1.51 |
ENSDART00000149581
|
ypel5
|
yippee-like 5 |
| chr19_+_7152966 | 1.49 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr19_+_42470396 | 1.46 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr2_+_54696042 | 1.46 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr18_-_226800 | 1.45 |
ENSDART00000165180
|
tarsl2
|
threonyl-tRNA synthetase-like 2 |
| chr3_-_10425313 | 1.45 |
ENSDART00000111833
|
pctp
|
phosphatidylcholine transfer protein |
| chr8_+_25247245 | 1.45 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr9_-_3149896 | 1.45 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr7_-_823495 | 1.42 |
ENSDART00000189926
ENSDART00000192318 |
si:cabz01076231.1
|
si:cabz01076231.1 |
| chr24_-_38644937 | 1.42 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr20_-_2949028 | 1.41 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr11_-_37411492 | 1.41 |
ENSDART00000166468
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr25_-_35113891 | 1.40 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr13_-_25284716 | 1.40 |
ENSDART00000039828
|
vcla
|
vinculin a |
| chr13_-_37519774 | 1.39 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr13_-_3516473 | 1.36 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr3_-_62380146 | 1.34 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr7_-_30082931 | 1.32 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr11_-_3959477 | 1.30 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr2_+_6181383 | 1.30 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr10_+_10677697 | 1.29 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr6_+_39184236 | 1.29 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr5_-_38342992 | 1.29 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr10_-_16028082 | 1.25 |
ENSDART00000122540
|
aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr23_+_22819971 | 1.25 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr21_-_20765338 | 1.24 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr9_-_35557397 | 1.20 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr1_-_9231952 | 1.18 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr11_+_41135055 | 1.18 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr2_-_5505094 | 1.18 |
ENSDART00000145035
|
saga
|
S-antigen; retina and pineal gland (arrestin) a |
| chr7_+_48999723 | 1.16 |
ENSDART00000182699
ENSDART00000166329 |
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr7_+_9922607 | 1.16 |
ENSDART00000184532
ENSDART00000113396 |
cers3a
|
ceramide synthase 3a |
| chr19_+_31183495 | 1.16 |
ENSDART00000088618
|
meox2b
|
mesenchyme homeobox 2b |
| chr19_+_33732188 | 1.16 |
ENSDART00000151192
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_-_11729 | 1.15 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr19_-_205104 | 1.15 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr16_-_31622777 | 1.15 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr21_-_36948 | 1.14 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr9_+_10014817 | 1.13 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr5_-_1999417 | 1.12 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr25_+_25123385 | 1.11 |
ENSDART00000163892
|
ldha
|
lactate dehydrogenase A4 |
| chr18_-_9046805 | 1.10 |
ENSDART00000134224
|
grm3
|
glutamate receptor, metabotropic 3 |
| chr10_+_21559605 | 1.08 |
ENSDART00000123648
ENSDART00000108584 |
pcdh1a3
pcdh1a3
|
protocadherin 1 alpha 3 protocadherin 1 alpha 3 |
| chr20_+_19212962 | 1.08 |
ENSDART00000063706
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr24_-_22702017 | 1.08 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr23_+_42131509 | 1.07 |
ENSDART00000184544
|
SHISAL2A
|
shisa like 2A |
| chr25_-_13871118 | 1.07 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr19_-_5103313 | 1.07 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr12_-_15620090 | 1.07 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr20_-_14665002 | 1.06 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr18_+_660578 | 1.06 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr19_+_42142381 | 1.06 |
ENSDART00000129915
|
kcnq4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr16_-_3127749 | 1.06 |
ENSDART00000063986
|
csf3r
|
colony stimulating factor 3 receptor (granulocyte) |
| chr11_-_43948885 | 1.03 |
ENSDART00000164522
|
CABZ01074203.1
|
|
| chr18_-_19103929 | 1.03 |
ENSDART00000188370
ENSDART00000177621 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr2_-_9489611 | 1.03 |
ENSDART00000146715
|
st6galnac3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr12_+_30586599 | 1.00 |
ENSDART00000124920
ENSDART00000126984 |
nrap
|
nebulin-related anchoring protein |
| chr23_+_39089574 | 0.98 |
ENSDART00000164711
|
nfatc2a
|
nuclear factor of activated T cells 2a |
| chr9_+_10014514 | 0.97 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr5_-_33935396 | 0.97 |
ENSDART00000133578
|
si:dkeyp-72a4.1
|
si:dkeyp-72a4.1 |
| chr3_-_28120092 | 0.96 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr18_+_19990412 | 0.96 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr1_+_7956030 | 0.95 |
ENSDART00000159655
|
CR855320.1
|
|
| chr24_-_38787457 | 0.95 |
ENSDART00000112318
ENSDART00000154461 |
ntn2
|
netrin 2 |
| chr10_+_575929 | 0.93 |
ENSDART00000129856
|
smad4a
|
SMAD family member 4a |
| chr6_-_24087279 | 0.91 |
ENSDART00000163445
|
zgc:109982
|
zgc:109982 |
| chr12_-_33359654 | 0.91 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_1378250 | 0.91 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr11_-_37880492 | 0.90 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr1_-_54420133 | 0.87 |
ENSDART00000190295
|
CABZ01043826.1
|
|
| chr13_+_31402067 | 0.85 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr10_-_105100 | 0.84 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr5_+_43470544 | 0.83 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr5_+_36932718 | 0.83 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr11_+_40032790 | 0.82 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr3_-_42981739 | 0.82 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr19_+_33732487 | 0.80 |
ENSDART00000010294
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr9_+_36314867 | 0.80 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr10_+_41159241 | 0.79 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr24_-_3342665 | 0.79 |
ENSDART00000167682
|
sugct
|
succinyl-CoA:glutarate-CoA transferase |
| chr17_+_10748366 | 0.79 |
ENSDART00000018683
ENSDART00000097274 |
ATG14
|
zgc:113944 |
| chr3_+_17744339 | 0.78 |
ENSDART00000132622
|
znf385c
|
zinc finger protein 385C |
| chr15_-_36533322 | 0.78 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr6_-_47953829 | 0.78 |
ENSDART00000157264
|
si:dkey-166d12.2
|
si:dkey-166d12.2 |
| chr12_+_33038757 | 0.77 |
ENSDART00000153146
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr24_+_12074340 | 0.77 |
ENSDART00000170997
ENSDART00000166227 |
ccr9b
|
chemokine (C-C motif) receptor 9b |
| chr2_-_48171112 | 0.76 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr22_-_5099824 | 0.76 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr18_+_14693682 | 0.75 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr12_-_3453589 | 0.75 |
ENSDART00000175918
|
CABZ01063170.1
|
|
| chr8_-_19649617 | 0.75 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr21_+_25054420 | 0.75 |
ENSDART00000065132
|
zgc:171740
|
zgc:171740 |
| chr11_-_15090564 | 0.75 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr17_-_24916889 | 0.75 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr3_-_40933415 | 0.74 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr19_-_6774871 | 0.74 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr19_+_43119698 | 0.74 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr18_+_6126506 | 0.74 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr24_-_12981423 | 0.74 |
ENSDART00000133166
ENSDART00000131370 ENSDART00000002992 |
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr6_-_57539141 | 0.73 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr19_-_47832853 | 0.73 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr13_-_280827 | 0.73 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr7_-_36358303 | 0.73 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr8_-_47329755 | 0.73 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr17_-_7440397 | 0.72 |
ENSDART00000162597
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr6_-_7842078 | 0.72 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr20_-_22378401 | 0.71 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr22_-_94352 | 0.70 |
ENSDART00000184883
|
ndufaf3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr16_+_48729947 | 0.70 |
ENSDART00000049051
|
brd2b
|
bromodomain containing 2b |
| chr16_-_13388821 | 0.70 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr21_-_41617372 | 0.69 |
ENSDART00000187171
|
BX005397.1
|
|
| chr12_+_30200652 | 0.69 |
ENSDART00000193405
ENSDART00000132335 |
ablim1b
|
actin binding LIM protein 1b |
| chr25_+_16945348 | 0.69 |
ENSDART00000016591
|
fgf6a
|
fibroblast growth factor 6a |
| chr7_-_36358735 | 0.68 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr1_-_30457062 | 0.68 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr5_-_26247215 | 0.68 |
ENSDART00000136806
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hey2_hey1+heyl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 1.1 | 3.3 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.8 | 3.9 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.8 | 3.0 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.6 | 3.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.6 | 2.5 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.5 | 1.4 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 4.0 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.4 | 4.3 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.4 | 1.4 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 | 2.5 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 2.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.3 | 1.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 1.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 1.4 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.2 | 10.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 0.5 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.2 | 1.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 2.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.7 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 1.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 1.6 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 1.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.2 | 0.6 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 1.6 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 1.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.4 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.1 | 0.4 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 1.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 5.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 1.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.8 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.9 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.8 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 1.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 17.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.3 | GO:0009085 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.5 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 1.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.6 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 2.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 2.1 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.8 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 1.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.2 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 2.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.7 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.8 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.4 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.3 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 3.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.7 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.1 | 1.9 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.5 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 1.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.8 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 2.3 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 2.6 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.1 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.0 | GO:0099622 | membrane repolarization during action potential(GO:0086011) cardiac muscle cell membrane repolarization(GO:0099622) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 2.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.7 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 2.5 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.4 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.6 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 2.3 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 3.2 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.5 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 4.8 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 1.4 | GO:0071774 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.9 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 4.6 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 2.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.0 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 1.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 3.9 | GO:0030428 | cell septum(GO:0030428) |
| 0.5 | 3.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 3.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.3 | 1.7 | GO:0071203 | WASH complex(GO:0071203) |
| 0.3 | 1.8 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 13.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.3 | 4.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 1.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 1.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 0.6 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.2 | 7.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 3.1 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.1 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 3.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 2.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.4 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 5.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 7.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.0 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.8 | 4.0 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.7 | 2.9 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.6 | 2.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 7.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.5 | 4.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 3.9 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.4 | 3.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.4 | 1.4 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.4 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.3 | 1.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.3 | 1.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 1.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 2.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 1.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 1.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 1.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.9 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 6.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 3.1 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 1.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 2.5 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.2 | 0.6 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.2 | 1.3 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 8.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 4.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 5.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.9 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.5 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 9.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.7 | GO:0099583 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.1 | 0.8 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.3 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 2.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.6 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.1 | 3.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.4 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 1.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 1.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 4.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 2.0 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.5 | GO:0005035 | death receptor activity(GO:0005035) |
| 0.1 | 2.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 6.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.3 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 3.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 2.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.6 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0052833 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 2.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.3 | 4.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 2.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 2.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 6.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 3.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 3.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 2.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 4.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 3.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |