Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
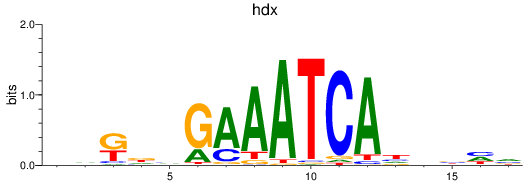
Results for hdx
Z-value: 0.48
Transcription factors associated with hdx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hdx
|
ENSDARG00000079382 | highly divergent homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hdx | dr11_v1_chr14_-_6225336_6225336 | -0.15 | 5.5e-01 | Click! |
Activity profile of hdx motif
Sorted Z-values of hdx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_34696503 | 0.53 |
ENSDART00000186106
|
CR339054.2
|
|
| chr5_-_30620625 | 0.52 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr22_-_278328 | 0.50 |
ENSDART00000098072
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr22_-_4778018 | 0.46 |
ENSDART00000143968
|
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr25_+_29160102 | 0.44 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr3_+_25154078 | 0.40 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr19_-_12324514 | 0.39 |
ENSDART00000122192
|
ncaldb
|
neurocalcin delta b |
| chr19_-_20270178 | 0.38 |
ENSDART00000144891
ENSDART00000090883 |
gpnmb
|
glycoprotein (transmembrane) nmb |
| chr11_-_29737088 | 0.36 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr23_-_4915118 | 0.35 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr3_+_35298078 | 0.30 |
ENSDART00000110126
|
cacng3b
|
calcium channel, voltage-dependent, gamma subunit 3b |
| chr4_+_28355058 | 0.30 |
ENSDART00000150799
|
si:ch73-263o4.3
|
si:ch73-263o4.3 |
| chr10_+_8101729 | 0.30 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr9_-_34871900 | 0.27 |
ENSDART00000026378
|
slc25a6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr10_+_39164638 | 0.26 |
ENSDART00000188997
|
CU633908.1
|
|
| chr13_+_33117528 | 0.26 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr23_+_26946429 | 0.26 |
ENSDART00000185564
|
cacnb3b
|
calcium channel, voltage-dependent, beta 3b |
| chr4_-_10599062 | 0.25 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr17_+_15433518 | 0.24 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr10_+_21563986 | 0.24 |
ENSDART00000100600
|
pcdh1a6
|
protocadherin 1 alpha 6 |
| chr4_+_7508316 | 0.24 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr14_-_30490763 | 0.24 |
ENSDART00000193166
ENSDART00000183471 ENSDART00000087859 |
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr4_-_69189894 | 0.24 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr9_-_46701326 | 0.23 |
ENSDART00000159971
|
si:ch73-193i22.1
|
si:ch73-193i22.1 |
| chr15_-_917274 | 0.23 |
ENSDART00000156624
|
si:dkey-77f5.15
|
si:dkey-77f5.15 |
| chr6_-_24143923 | 0.23 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr7_-_28413224 | 0.22 |
ENSDART00000076502
|
rerglb
|
RERG/RAS-like b |
| chr23_+_5108374 | 0.22 |
ENSDART00000114263
|
zgc:194242
|
zgc:194242 |
| chr14_-_2213660 | 0.21 |
ENSDART00000162537
|
pcdh2ab3
|
protocadherin 2 alpha b 3 |
| chr22_-_8006342 | 0.21 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr23_+_17437554 | 0.21 |
ENSDART00000184282
|
BX649300.3
|
|
| chr18_-_8885792 | 0.21 |
ENSDART00000143619
|
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr16_+_48753664 | 0.21 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr21_-_43952958 | 0.20 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr2_-_40199780 | 0.20 |
ENSDART00000113901
|
ccl34a.4
|
chemokine (C-C motif) ligand 34a, duplicate 4 |
| chr18_+_808911 | 0.20 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr1_-_8928841 | 0.20 |
ENSDART00000103652
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr17_+_15882533 | 0.20 |
ENSDART00000164124
|
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr5_-_30535327 | 0.19 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr24_-_14212521 | 0.19 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr17_-_2834764 | 0.19 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr4_-_8040436 | 0.19 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr21_+_39336285 | 0.18 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr25_-_225964 | 0.18 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr20_-_46554440 | 0.18 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr23_-_31633201 | 0.18 |
ENSDART00000143335
ENSDART00000053531 |
slc2a12
|
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr16_+_25196572 | 0.17 |
ENSDART00000141956
|
tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr13_+_33688474 | 0.16 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr18_+_9810642 | 0.16 |
ENSDART00000143965
|
CACNA2D1
|
si:dkey-266j7.2 |
| chr13_+_40686133 | 0.16 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr8_-_21268303 | 0.16 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr17_+_44780166 | 0.16 |
ENSDART00000156260
|
tmem63c
|
transmembrane protein 63C |
| chr8_+_2773776 | 0.16 |
ENSDART00000156796
ENSDART00000154529 ENSDART00000155092 |
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr10_-_36793412 | 0.16 |
ENSDART00000185966
|
dhrs13a.2
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 2 |
| chr8_-_38965260 | 0.15 |
ENSDART00000181495
|
CR392001.3
|
|
| chr18_-_17077419 | 0.15 |
ENSDART00000148714
|
il17c
|
interleukin 17c |
| chr15_+_31428332 | 0.15 |
ENSDART00000141429
|
or103-5
|
odorant receptor, family C, subfamily 103, member 5 |
| chr11_+_8152872 | 0.15 |
ENSDART00000091638
ENSDART00000138057 ENSDART00000166379 |
samd13
|
sterile alpha motif domain containing 13 |
| chr13_+_22675802 | 0.14 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr16_+_34523515 | 0.14 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr2_+_45484183 | 0.14 |
ENSDART00000183490
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr16_-_33930060 | 0.14 |
ENSDART00000110743
ENSDART00000101898 |
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr10_+_21588002 | 0.14 |
ENSDART00000100596
|
pcdh1a6
|
protocadherin 1 alpha 6 |
| chr2_-_36925561 | 0.13 |
ENSDART00000187690
|
map1sb
|
microtubule-associated protein 1Sb |
| chr3_-_15264698 | 0.13 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr7_+_49865049 | 0.13 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr13_-_9895564 | 0.13 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr9_-_16877456 | 0.13 |
ENSDART00000161105
ENSDART00000160869 |
fbxl3a
|
F-box and leucine-rich repeat protein 3a |
| chr21_+_11244068 | 0.13 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr8_+_8468139 | 0.13 |
ENSDART00000186684
|
comta
|
catechol-O-methyltransferase a |
| chr25_-_4313699 | 0.12 |
ENSDART00000154038
|
syt7a
|
synaptotagmin VIIa |
| chr18_-_14691727 | 0.12 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr7_+_58699718 | 0.12 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr15_-_6946286 | 0.12 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr2_-_14390627 | 0.12 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr2_+_45696743 | 0.12 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
|
| chr10_+_36155366 | 0.12 |
ENSDART00000165466
|
waif2
|
Wnt-activated inhibitory factor 2 |
| chr14_+_15620640 | 0.12 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr21_-_7940043 | 0.12 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr21_-_28439596 | 0.12 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr14_-_41678749 | 0.12 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor like 1b |
| chr6_-_41021571 | 0.12 |
ENSDART00000103776
|
hdhd3
|
haloacid dehalogenase-like hydrolase domain containing 3 |
| chr8_+_12975963 | 0.11 |
ENSDART00000132821
ENSDART00000181755 ENSDART00000185061 ENSDART00000193727 |
dram2a
|
DNA-damage regulated autophagy modulator 2a |
| chr6_-_2133737 | 0.11 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr19_+_23298037 | 0.11 |
ENSDART00000183948
|
irgf1
|
immunity-related GTPase family, f1 |
| chr25_+_10811551 | 0.11 |
ENSDART00000167730
|
anpepb
|
alanyl (membrane) aminopeptidase b |
| chr5_-_33825465 | 0.11 |
ENSDART00000110645
|
dab2ipb
|
DAB2 interacting protein b |
| chr25_+_2282265 | 0.11 |
ENSDART00000076417
|
cdkn1bb
|
cyclin-dependent kinase inhibitor 1Bb |
| chr16_-_31933740 | 0.10 |
ENSDART00000125411
|
si:ch1073-90m23.1
|
si:ch1073-90m23.1 |
| chr5_+_27267186 | 0.10 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr3_+_16724614 | 0.10 |
ENSDART00000182135
|
gys1
|
glycogen synthase 1 (muscle) |
| chr14_+_30398546 | 0.10 |
ENSDART00000053925
|
mtmr7a
|
myotubularin related protein 7a |
| chr21_-_21089781 | 0.10 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr4_+_2269815 | 0.10 |
ENSDART00000075788
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr8_+_7033049 | 0.10 |
ENSDART00000064172
ENSDART00000134440 |
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr19_+_2685779 | 0.10 |
ENSDART00000160533
ENSDART00000097531 |
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr15_+_42235449 | 0.10 |
ENSDART00000114801
ENSDART00000182053 |
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr25_+_15997957 | 0.10 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr14_-_30490465 | 0.10 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr6_+_18569453 | 0.10 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr14_-_25599002 | 0.09 |
ENSDART00000040955
|
slc25a48
|
solute carrier family 25, member 48 |
| chr2_+_56463167 | 0.09 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr10_-_18492617 | 0.09 |
ENSDART00000179936
ENSDART00000193799 ENSDART00000193198 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr6_+_49082796 | 0.09 |
ENSDART00000182446
|
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr20_-_25542183 | 0.09 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr3_+_33918510 | 0.09 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr16_+_45571956 | 0.09 |
ENSDART00000143867
|
syngap1b
|
synaptic Ras GTPase activating protein 1b |
| chr1_+_27867706 | 0.09 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr19_+_41173386 | 0.09 |
ENSDART00000142773
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr8_+_53120278 | 0.09 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr18_+_14477740 | 0.09 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr23_-_44226556 | 0.09 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr12_-_44009049 | 0.09 |
ENSDART00000191511
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr1_-_23157583 | 0.09 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr14_-_2202652 | 0.08 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr4_+_41227127 | 0.08 |
ENSDART00000136445
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr20_+_34792166 | 0.08 |
ENSDART00000061523
|
il17a/f3
|
interleukin 17a/f3 |
| chr20_-_35052823 | 0.08 |
ENSDART00000153033
|
kif26bb
|
kinesin family member 26Bb |
| chr18_-_6742287 | 0.08 |
ENSDART00000140752
|
miox
|
myo-inositol oxygenase |
| chr9_+_22080122 | 0.08 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr6_+_72040 | 0.08 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr3_-_25492361 | 0.08 |
ENSDART00000147322
ENSDART00000055473 |
grb2b
|
growth factor receptor-bound protein 2b |
| chr7_-_8052829 | 0.08 |
ENSDART00000172931
|
si:ch211-163c2.2
|
si:ch211-163c2.2 |
| chr15_-_8177919 | 0.08 |
ENSDART00000101463
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr7_+_65387374 | 0.08 |
ENSDART00000188680
|
CLEC3A
|
C-type lectin domain family 3 member A |
| chr4_+_77060861 | 0.08 |
ENSDART00000174271
ENSDART00000174393 ENSDART00000150450 |
si:dkey-240n22.8
|
si:dkey-240n22.8 |
| chr8_-_36370552 | 0.07 |
ENSDART00000097932
ENSDART00000148323 |
si:busm1-104n07.3
|
si:busm1-104n07.3 |
| chr14_-_2352384 | 0.07 |
ENSDART00000170666
|
si:ch73-233f7.7
|
si:ch73-233f7.7 |
| chr17_-_23674495 | 0.07 |
ENSDART00000122209
|
ptena
|
phosphatase and tensin homolog A |
| chr18_-_19491162 | 0.07 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr12_-_26620728 | 0.07 |
ENSDART00000087067
|
hexdc
|
hexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing |
| chr2_+_34767171 | 0.07 |
ENSDART00000145451
|
astn1
|
astrotactin 1 |
| chr12_+_29173523 | 0.07 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr19_-_18626952 | 0.06 |
ENSDART00000168004
ENSDART00000162034 ENSDART00000165486 ENSDART00000167971 |
rps18
|
ribosomal protein S18 |
| chr10_-_25570647 | 0.06 |
ENSDART00000134215
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr20_+_41756996 | 0.06 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr20_-_26841544 | 0.06 |
ENSDART00000088875
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr25_-_21609645 | 0.06 |
ENSDART00000087817
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr18_-_17077596 | 0.06 |
ENSDART00000111821
|
il17c
|
interleukin 17c |
| chr5_+_27421025 | 0.06 |
ENSDART00000098590
|
cyb561a3a
|
cytochrome b561 family, member A3a |
| chr10_+_21722892 | 0.06 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr19_-_3488860 | 0.06 |
ENSDART00000172520
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr1_-_11519934 | 0.06 |
ENSDART00000162060
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr5_+_10046643 | 0.06 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr20_+_2039518 | 0.06 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr16_+_38820486 | 0.06 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr18_-_7112882 | 0.06 |
ENSDART00000176292
|
si:dkey-88e18.8
|
si:dkey-88e18.8 |
| chr20_+_32552912 | 0.06 |
ENSDART00000009691
|
scml4
|
Scm polycomb group protein like 4 |
| chr5_+_62356304 | 0.05 |
ENSDART00000148381
|
aspa
|
aspartoacylase |
| chr25_-_18778356 | 0.05 |
ENSDART00000111425
|
fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr7_-_35380186 | 0.05 |
ENSDART00000148430
|
slc6a2
|
solute carrier family 6 (neurotransmitter transporter), member 2 |
| chr20_+_19562383 | 0.05 |
ENSDART00000103798
|
nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr10_-_32663759 | 0.05 |
ENSDART00000126727
|
atg101
|
autophagy related 101 |
| chr11_-_25775447 | 0.05 |
ENSDART00000161301
|
plch2b
|
phospholipase C, eta 2b |
| chr18_-_16392448 | 0.05 |
ENSDART00000180253
|
mgat4c
|
mgat4 family, member C |
| chr9_+_32301017 | 0.05 |
ENSDART00000127916
ENSDART00000183298 ENSDART00000143103 |
hspe1
|
heat shock 10 protein 1 |
| chr10_-_36808348 | 0.05 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr21_-_2322102 | 0.05 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr20_+_2240278 | 0.05 |
ENSDART00000041250
|
tmem200a
|
transmembrane protein 200A |
| chr16_-_54471235 | 0.05 |
ENSDART00000061572
|
cyp11c1
|
cytochrome P450, family 11, subfamily C, polypeptide 1 |
| chr5_-_38342992 | 0.05 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr1_-_9104631 | 0.05 |
ENSDART00000146642
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr16_+_19029297 | 0.05 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr21_+_39365920 | 0.05 |
ENSDART00000140940
|
cluhb
|
clustered mitochondria (cluA/CLU1) homolog b |
| chr22_+_10201826 | 0.05 |
ENSDART00000006513
ENSDART00000132641 |
pdhb
|
pyruvate dehydrogenase E1 beta subunit |
| chr21_+_43882274 | 0.04 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr8_-_16592491 | 0.04 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr16_-_21785261 | 0.04 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr4_-_13156971 | 0.04 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr16_+_51154918 | 0.04 |
ENSDART00000058290
|
dhdds
|
dehydrodolichyl diphosphate synthase |
| chr1_-_25177086 | 0.04 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr17_-_20143946 | 0.04 |
ENSDART00000138911
|
actn2b
|
actinin, alpha 2b |
| chr17_+_6535694 | 0.04 |
ENSDART00000191681
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr1_+_41588170 | 0.04 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr2_-_15563194 | 0.04 |
ENSDART00000049589
|
col11a1b
|
collagen, type XI, alpha 1b |
| chr9_-_11551608 | 0.04 |
ENSDART00000128629
|
fev
|
FEV (ETS oncogene family) |
| chr25_+_23489979 | 0.04 |
ENSDART00000158200
|
CU024870.1
|
|
| chr21_+_21679086 | 0.04 |
ENSDART00000146225
|
or125-5
|
odorant receptor, family E, subfamily 125, member 5 |
| chr3_-_40945710 | 0.04 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr16_+_16933002 | 0.04 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr6_-_31323984 | 0.04 |
ENSDART00000183660
ENSDART00000184146 ENSDART00000180156 |
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr7_+_38962459 | 0.04 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr6_-_10305918 | 0.04 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr10_-_11203164 | 0.04 |
ENSDART00000040308
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr5_+_13373593 | 0.04 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr21_-_32684570 | 0.04 |
ENSDART00000162873
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr16_+_32729223 | 0.04 |
ENSDART00000125663
|
si:dkey-165n16.1
|
si:dkey-165n16.1 |
| chr9_+_8364553 | 0.04 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr16_+_6632969 | 0.04 |
ENSDART00000109276
|
sall3a
|
spalt-like transcription factor 3a |
| chr5_+_57210237 | 0.04 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr5_-_3885727 | 0.04 |
ENSDART00000143250
|
mlxipl
|
MLX interacting protein like |
| chr21_-_35534401 | 0.04 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr25_+_3564220 | 0.04 |
ENSDART00000170005
|
zgc:194202
|
zgc:194202 |
| chr12_-_11294979 | 0.04 |
ENSDART00000148850
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr5_+_25680845 | 0.04 |
ENSDART00000139701
ENSDART00000009952 |
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr6_-_40573221 | 0.04 |
ENSDART00000157203
ENSDART00000154951 |
tomm6
|
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr25_+_36333993 | 0.04 |
ENSDART00000184379
|
CR354435.2
|
|
| chr1_-_52431220 | 0.04 |
ENSDART00000111256
|
zgc:194101
|
zgc:194101 |
| chr8_-_33568964 | 0.04 |
ENSDART00000138921
|
mvb12bb
|
multivesicular body subunit 12Bb |
| chr18_+_35742838 | 0.04 |
ENSDART00000088504
ENSDART00000140386 |
rasgrp4
|
RAS guanyl releasing protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hdx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.3 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.2 | GO:0071634 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.2 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.2 | GO:0046323 | glucose import(GO:0046323) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.1 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.2 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0005330 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |