Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
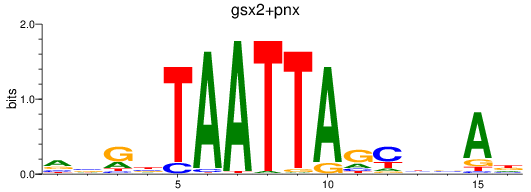
Results for gsx2+pnx
Z-value: 1.01
Transcription factors associated with gsx2+pnx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pnx
|
ENSDARG00000025899 | posterior neuron-specific homeobox |
|
gsx2
|
ENSDARG00000043322 | GS homeobox 2 |
|
gsx2
|
ENSDARG00000116417 | GS homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsx2 | dr11_v1_chr20_-_22484621_22484621 | -0.72 | 5.6e-04 | Click! |
| pnx | dr11_v1_chr10_+_2799285_2799285 | -0.67 | 1.7e-03 | Click! |
Activity profile of gsx2+pnx motif
Sorted Z-values of gsx2+pnx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22310919 | 7.22 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr3_-_50443607 | 3.62 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr17_-_37395460 | 3.46 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr22_+_7738966 | 3.27 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr11_+_38280454 | 3.27 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr11_+_43043171 | 3.22 |
ENSDART00000180344
|
CABZ01092982.1
|
|
| chr9_-_14683574 | 3.10 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr5_+_51597677 | 3.05 |
ENSDART00000048210
ENSDART00000184797 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr6_+_40354424 | 2.90 |
ENSDART00000047416
|
slc4a8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr8_-_21372446 | 2.77 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr25_-_36827491 | 2.70 |
ENSDART00000170941
|
CU929365.1
|
|
| chr25_+_29160102 | 2.66 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr25_+_31277415 | 2.60 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr6_-_39344259 | 2.58 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr10_-_44508249 | 2.53 |
ENSDART00000160018
|
DUSP26
|
dual specificity phosphatase 26 |
| chr7_-_71758307 | 2.49 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr18_+_9615147 | 2.49 |
ENSDART00000160284
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr24_+_3307857 | 2.44 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr23_+_40460333 | 2.38 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr8_+_53423408 | 2.33 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr3_-_61218272 | 2.30 |
ENSDART00000133091
ENSDART00000055068 |
pvalb5
|
parvalbumin 5 |
| chr19_+_1831911 | 2.19 |
ENSDART00000166653
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr3_-_61217753 | 2.15 |
ENSDART00000182630
|
pvalb5
|
parvalbumin 5 |
| chr11_-_472547 | 2.13 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr1_-_50859053 | 2.10 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr19_+_9295244 | 2.05 |
ENSDART00000132255
ENSDART00000144299 |
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr21_-_37790727 | 2.05 |
ENSDART00000162907
|
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr3_-_60827402 | 2.03 |
ENSDART00000053494
|
anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr8_-_23780334 | 2.03 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr21_+_3244146 | 1.99 |
ENSDART00000127740
|
ctif
|
CBP80/20-dependent translation initiation factor |
| chr22_-_21897203 | 1.99 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr18_+_5137241 | 1.94 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr3_-_30625219 | 1.85 |
ENSDART00000151698
|
syt3
|
synaptotagmin III |
| chr19_-_657439 | 1.78 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr16_-_13818061 | 1.77 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr18_+_11506561 | 1.76 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr4_+_21129752 | 1.75 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr25_+_31276842 | 1.74 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr15_+_44366556 | 1.73 |
ENSDART00000133449
|
GUCY1A2
|
guanylate cyclase 1 soluble subunit alpha 2 |
| chr21_+_1381276 | 1.71 |
ENSDART00000192907
|
TCF4
|
transcription factor 4 |
| chr2_+_55982300 | 1.69 |
ENSDART00000183903
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr21_-_42097736 | 1.69 |
ENSDART00000100000
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr13_+_15190677 | 1.65 |
ENSDART00000142240
ENSDART00000129045 |
mavs
|
mitochondrial antiviral signaling protein |
| chr9_-_6372535 | 1.61 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr20_-_1378514 | 1.59 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr1_+_14283692 | 1.58 |
ENSDART00000017679
|
ppp2r2ca
|
protein phosphatase 2, regulatory subunit B, gamma a |
| chr7_-_27685365 | 1.55 |
ENSDART00000188342
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr12_+_47446158 | 1.54 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr2_+_56937548 | 1.48 |
ENSDART00000189308
|
CABZ01117752.1
|
|
| chr24_-_41320037 | 1.47 |
ENSDART00000129058
|
rheb
|
Ras homolog, mTORC1 binding |
| chr20_+_35382482 | 1.45 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr5_-_30074332 | 1.44 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr23_+_28582865 | 1.44 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr22_+_17205608 | 1.44 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr11_+_26476153 | 1.43 |
ENSDART00000103507
|
unm_sa1614
|
un-named sa1614 |
| chr7_+_54642005 | 1.42 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr18_+_8833251 | 1.42 |
ENSDART00000143519
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr24_-_33276139 | 1.41 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr6_+_3280939 | 1.40 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr7_+_65261576 | 1.39 |
ENSDART00000169566
|
bco1
|
beta-carotene oxygenase 1 |
| chr19_-_5805923 | 1.39 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr15_-_16098531 | 1.38 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr3_-_34084387 | 1.34 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr2_-_28671139 | 1.33 |
ENSDART00000165272
ENSDART00000164657 |
dhcr7
|
7-dehydrocholesterol reductase |
| chr8_+_14855471 | 1.33 |
ENSDART00000134775
|
si:dkey-21p1.3
|
si:dkey-21p1.3 |
| chr6_+_34511886 | 1.33 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr16_+_13818500 | 1.31 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr3_-_61099004 | 1.30 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr4_+_76575585 | 1.29 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr9_-_813511 | 1.28 |
ENSDART00000082368
|
marco
|
macrophage receptor with collagenous structure |
| chr12_+_45677293 | 1.27 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr15_-_12011202 | 1.24 |
ENSDART00000160427
ENSDART00000168715 |
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr18_+_10929661 | 1.22 |
ENSDART00000061838
ENSDART00000113146 |
ttc38
|
tetratricopeptide repeat domain 38 |
| chr6_-_30210378 | 1.22 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr4_+_9450415 | 1.21 |
ENSDART00000189690
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr15_-_33925851 | 1.21 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr2_+_50608099 | 1.20 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr15_-_38154616 | 1.20 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr9_-_22834860 | 1.19 |
ENSDART00000146486
|
neb
|
nebulin |
| chr21_+_45841731 | 1.19 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr23_+_4646194 | 1.16 |
ENSDART00000092344
|
LO017700.1
|
|
| chr10_+_35952532 | 1.16 |
ENSDART00000184730
|
rtn4rl1a
|
reticulon 4 receptor-like 1a |
| chr18_-_43884044 | 1.16 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr7_+_67486807 | 1.14 |
ENSDART00000159989
|
cpne7
|
copine VII |
| chr18_+_3332999 | 1.14 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr23_-_1348933 | 1.14 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr1_-_14332283 | 1.14 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr15_-_36533322 | 1.14 |
ENSDART00000156466
ENSDART00000121755 |
si:dkey-262k9.4
|
si:dkey-262k9.4 |
| chr13_-_44630111 | 1.14 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr2_-_1569250 | 1.14 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr24_-_31425799 | 1.13 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr8_+_52637507 | 1.13 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr11_-_2131280 | 1.13 |
ENSDART00000008409
|
calcoco1b
|
calcium binding and coiled-coil domain 1b |
| chr22_+_28337429 | 1.12 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr19_-_2861444 | 1.12 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr10_+_3049636 | 1.12 |
ENSDART00000081794
ENSDART00000183167 ENSDART00000191634 ENSDART00000183514 |
rasgrf2a
|
Ras protein-specific guanine nucleotide-releasing factor 2a |
| chr3_-_23407720 | 1.11 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr24_-_24282427 | 1.11 |
ENSDART00000123299
|
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr22_+_2254972 | 1.11 |
ENSDART00000144906
|
znf1157
|
zinc finger protein 1157 |
| chr19_-_5103313 | 1.10 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr15_-_12011390 | 1.09 |
ENSDART00000187403
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr25_+_31267268 | 1.09 |
ENSDART00000181239
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr24_-_29586082 | 1.08 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr18_-_2433011 | 1.04 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr14_-_9713549 | 1.01 |
ENSDART00000193356
ENSDART00000166739 |
si:zfos-2326c3.2
|
si:zfos-2326c3.2 |
| chr21_+_6751405 | 1.01 |
ENSDART00000037265
ENSDART00000146371 |
olfm1b
|
olfactomedin 1b |
| chr6_-_48473694 | 1.01 |
ENSDART00000154237
|
ppm1j
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr1_+_55755304 | 0.99 |
ENSDART00000144983
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr2_-_57076687 | 0.99 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr20_-_46362606 | 0.99 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr6_+_39279104 | 0.98 |
ENSDART00000023686
|
ankrd33ab
|
ankyrin repeat domain 33Ab |
| chr9_-_950004 | 0.98 |
ENSDART00000168917
|
SCTR
|
secretin receptor |
| chr23_-_44219902 | 0.98 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr21_-_20939488 | 0.97 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr7_-_24046999 | 0.97 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr10_+_42690374 | 0.97 |
ENSDART00000123496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr10_-_2524917 | 0.97 |
ENSDART00000188642
|
CU856539.1
|
|
| chr5_+_30624183 | 0.97 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr14_-_4145594 | 0.96 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr11_+_45323455 | 0.96 |
ENSDART00000189249
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr1_-_59287410 | 0.96 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr20_-_14925281 | 0.93 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr18_+_1154189 | 0.93 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr15_+_34592215 | 0.93 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr2_+_6963296 | 0.91 |
ENSDART00000147146
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr19_-_46091497 | 0.90 |
ENSDART00000178772
ENSDART00000167255 |
ptdss1b
si:dkey-108k24.2
|
phosphatidylserine synthase 1b si:dkey-108k24.2 |
| chr12_-_4532066 | 0.90 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr17_+_3379673 | 0.89 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr20_-_1635922 | 0.89 |
ENSDART00000181502
|
CR846082.1
|
|
| chr11_+_7264457 | 0.87 |
ENSDART00000154182
|
reep6
|
receptor accessory protein 6 |
| chr8_-_45277370 | 0.87 |
ENSDART00000146364
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr10_-_5581487 | 0.86 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr6_+_6924637 | 0.86 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr22_-_8388678 | 0.86 |
ENSDART00000184389
|
CABZ01061498.1
|
|
| chr1_-_58963395 | 0.86 |
ENSDART00000130415
|
CABZ01115881.1
|
|
| chr19_+_5543072 | 0.84 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr17_-_37195354 | 0.83 |
ENSDART00000190963
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr2_-_14390627 | 0.82 |
ENSDART00000172367
|
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr4_+_9400012 | 0.82 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr8_+_52503340 | 0.81 |
ENSDART00000168838
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr19_-_5103141 | 0.81 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_-_49908978 | 0.80 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr16_+_31804590 | 0.79 |
ENSDART00000167321
|
wnt4b
|
wingless-type MMTV integration site family, member 4b |
| chr23_-_46020226 | 0.79 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr8_+_7756893 | 0.78 |
ENSDART00000191894
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr15_+_19646902 | 0.78 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr8_+_36942262 | 0.78 |
ENSDART00000188173
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr21_-_5205617 | 0.76 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr17_+_20174812 | 0.76 |
ENSDART00000186628
|
si:ch211-248a14.8
|
si:ch211-248a14.8 |
| chr7_+_30787903 | 0.76 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr1_+_52392511 | 0.76 |
ENSDART00000144025
|
si:ch211-217k17.8
|
si:ch211-217k17.8 |
| chr20_-_13774826 | 0.75 |
ENSDART00000063831
|
opn8c
|
opsin 8, group member c |
| chr24_-_36680261 | 0.75 |
ENSDART00000059507
|
ccr10
|
chemokine (C-C motif) receptor 10 |
| chr11_-_42554290 | 0.75 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr14_-_33824329 | 0.74 |
ENSDART00000189271
ENSDART00000180458 |
vimr2
|
vimentin-related 2 |
| chr16_-_25568512 | 0.74 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr25_+_2361721 | 0.74 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr3_+_13637383 | 0.74 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr18_-_2727764 | 0.74 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr19_-_22541652 | 0.73 |
ENSDART00000188208
|
pleca
|
plectin a |
| chr22_-_30770751 | 0.73 |
ENSDART00000172115
|
AL831726.2
|
|
| chr15_-_22074315 | 0.73 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr8_+_20776654 | 0.73 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr15_-_2726662 | 0.72 |
ENSDART00000108856
|
si:ch211-235m3.5
|
si:ch211-235m3.5 |
| chr19_-_44064842 | 0.72 |
ENSDART00000151541
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr4_+_73085993 | 0.72 |
ENSDART00000165749
|
si:ch73-170d6.2
|
si:ch73-170d6.2 |
| chr9_-_5263947 | 0.71 |
ENSDART00000088342
|
cytip
|
cytohesin 1 interacting protein |
| chr16_+_45424907 | 0.71 |
ENSDART00000134471
|
phf1
|
PHD finger protein 1 |
| chr12_-_46112892 | 0.71 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr2_+_33382648 | 0.70 |
ENSDART00000137207
ENSDART00000098831 |
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr19_-_22541284 | 0.70 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr23_-_1658980 | 0.70 |
ENSDART00000186227
|
CU693481.1
|
|
| chr6_+_24420523 | 0.70 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr25_-_13839743 | 0.70 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr5_+_19343880 | 0.68 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr2_+_8779164 | 0.68 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr5_+_69747417 | 0.67 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr12_+_30265802 | 0.67 |
ENSDART00000182412
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr10_+_37137464 | 0.66 |
ENSDART00000114909
|
cuedc1a
|
CUE domain containing 1a |
| chr10_+_16188761 | 0.66 |
ENSDART00000193244
|
ctxn3
|
cortexin 3 |
| chr13_-_25719628 | 0.66 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr20_-_18382708 | 0.64 |
ENSDART00000170864
ENSDART00000166762 ENSDART00000191333 |
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr19_-_31402429 | 0.64 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr17_+_20204122 | 0.64 |
ENSDART00000078672
|
gnrh3
|
gonadotropin-releasing hormone 3 |
| chr3_+_34919810 | 0.64 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr6_-_30954268 | 0.64 |
ENSDART00000154523
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr7_+_18106727 | 0.63 |
ENSDART00000190307
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr4_-_5019113 | 0.63 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr25_-_4713461 | 0.62 |
ENSDART00000155302
|
drd4a
|
dopamine receptor D4a |
| chr10_-_5847655 | 0.62 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr5_-_38063585 | 0.62 |
ENSDART00000164947
|
si:dkey-111e8.5
|
si:dkey-111e8.5 |
| chr15_-_14552101 | 0.61 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr2_+_30369116 | 0.60 |
ENSDART00000142137
|
pi15b
|
peptidase inhibitor 15b |
| chr10_-_17083180 | 0.60 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr12_-_33354409 | 0.60 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_6751760 | 0.59 |
ENSDART00000135914
|
olfm1b
|
olfactomedin 1b |
| chr18_+_19008560 | 0.59 |
ENSDART00000184217
ENSDART00000060735 |
slc24a1
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 |
| chr11_+_2506516 | 0.59 |
ENSDART00000130886
ENSDART00000189767 |
NABP2
|
si:ch73-190f16.2 |
| chr16_-_41936932 | 0.59 |
ENSDART00000111956
|
gramd1a
|
GRAM domain containing 1A |
| chr10_+_21867307 | 0.58 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsx2+pnx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.8 | 3.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.5 | 1.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.5 | 1.9 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 1.7 | GO:0098586 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.3 | 3.6 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.3 | 2.8 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.3 | 0.9 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.3 | 0.9 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.2 | 2.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 3.1 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.6 | GO:0070207 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.2 | 0.7 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.2 | 0.8 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 1.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.6 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 1.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.2 | 0.7 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 0.7 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 0.7 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 0.7 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.5 | GO:2000374 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.2 | 0.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 2.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.2 | 1.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.2 | 3.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 1.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.9 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.4 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 2.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 3.6 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 5.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 1.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.6 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.3 | GO:0099590 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 8.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 4.0 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.3 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 1.3 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 1.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 2.4 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 1.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.8 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 2.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.0 | 0.8 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 3.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.7 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 1.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 2.0 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 1.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.8 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.0 | 2.4 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.7 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 1.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.1 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 2.3 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.9 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.8 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 1.8 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 2.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 1.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 1.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 5.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.8 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 3.7 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.5 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 4.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 5.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.2 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.7 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 15.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.5 | 1.9 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 2.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.0 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.3 | 2.8 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.3 | 1.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.7 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 3.1 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 1.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.2 | 2.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 2.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 0.6 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.2 | 2.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.7 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.5 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 0.8 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 1.7 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.7 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 2.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 8.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.1 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 2.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 3.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.7 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.6 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.0 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 2.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 6.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.5 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 2.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.7 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.7 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 1.0 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |