Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
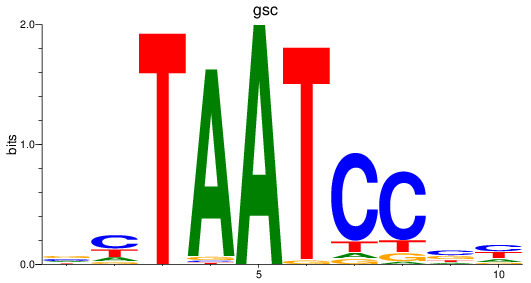
Results for gsc
Z-value: 1.48
Transcription factors associated with gsc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gsc
|
ENSDARG00000059073 | goosecoid |
|
gsc
|
ENSDARG00000111184 | goosecoid |
|
gsc
|
ENSDARG00000115937 | goosecoid |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gsc | dr11_v1_chr17_-_19344999_19344999 | 0.71 | 6.7e-04 | Click! |
Activity profile of gsc motif
Sorted Z-values of gsc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_54077740 | 14.28 |
ENSDART00000027000
|
rho
|
rhodopsin |
| chr17_-_51202339 | 8.70 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr14_+_32838110 | 7.37 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr6_+_41186320 | 7.24 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr6_-_345503 | 6.48 |
ENSDART00000168901
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr11_-_15090118 | 6.25 |
ENSDART00000171118
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr3_-_32170850 | 6.15 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr11_-_15090564 | 5.55 |
ENSDART00000162079
|
slc1a8a
|
solute carrier family 1 (glutamate transporter), member 8a |
| chr14_+_32839535 | 5.22 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr15_+_45640906 | 5.10 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr19_+_40856534 | 5.07 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr14_+_32837914 | 5.05 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr7_-_13882988 | 4.76 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr10_-_20453995 | 4.62 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr19_+_40856807 | 4.60 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr14_+_46313396 | 4.46 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr5_+_28973264 | 4.28 |
ENSDART00000005638
|
stxbp1b
|
syntaxin binding protein 1b |
| chr1_-_58009216 | 4.14 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr5_+_28972935 | 4.12 |
ENSDART00000193274
|
stxbp1b
|
syntaxin binding protein 1b |
| chr23_+_39854566 | 3.99 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr2_+_7192966 | 3.90 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr14_+_46313135 | 3.86 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr22_+_34701848 | 3.83 |
ENSDART00000082066
|
atpv0e2
|
ATPase H+ transporting V0 subunit e2 |
| chr5_-_38161033 | 3.54 |
ENSDART00000145907
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr6_+_48618512 | 3.49 |
ENSDART00000111190
|
FAM19A3
|
si:dkey-238f9.1 |
| chr3_-_28828242 | 3.41 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr17_-_16965809 | 3.34 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr14_-_36378494 | 3.12 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr17_+_36627099 | 2.99 |
ENSDART00000154104
|
impg1b
|
interphotoreceptor matrix proteoglycan 1b |
| chr10_-_22845485 | 2.92 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr21_-_28235361 | 2.90 |
ENSDART00000164458
|
nrxn2a
|
neurexin 2a |
| chr13_+_29462249 | 2.87 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr7_+_56615554 | 2.77 |
ENSDART00000098430
|
dpep1
|
dipeptidase 1 |
| chr2_-_5356686 | 2.75 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr12_+_5530247 | 2.67 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr10_-_22249444 | 2.64 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr9_+_33357011 | 2.57 |
ENSDART00000088569
|
nyx
|
nyctalopin |
| chr23_+_6232895 | 2.57 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr2_-_4797512 | 2.56 |
ENSDART00000160765
|
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr16_+_32059785 | 2.53 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr13_+_3954715 | 2.50 |
ENSDART00000182477
ENSDART00000192142 ENSDART00000190962 |
lrrc73
|
leucine rich repeat containing 73 |
| chr8_+_23142946 | 2.50 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr3_+_34919810 | 2.50 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr13_+_3954540 | 2.50 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr5_+_1278092 | 2.48 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr7_-_49594995 | 2.40 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr15_+_9053059 | 2.40 |
ENSDART00000012039
|
ppm1na
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Na (putative) |
| chr23_-_21797517 | 2.40 |
ENSDART00000110041
|
lrrc38a
|
leucine rich repeat containing 38a |
| chr13_+_12528043 | 2.38 |
ENSDART00000057761
|
rrh
|
retinal pigment epithelium-derived rhodopsin homolog |
| chr16_-_17188294 | 2.37 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr17_-_15149192 | 2.35 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr2_+_24199073 | 2.34 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr2_-_30784198 | 2.34 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr22_+_5103349 | 2.34 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr25_+_27923846 | 2.32 |
ENSDART00000047007
|
slc13a1
|
solute carrier family 13 member 1 |
| chr19_+_1097393 | 2.28 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr1_+_41690402 | 2.22 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr19_-_10373630 | 2.21 |
ENSDART00000131517
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr9_+_13685921 | 2.18 |
ENSDART00000145775
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr1_-_45920632 | 2.14 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr24_-_6501211 | 2.14 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr11_-_44873780 | 2.10 |
ENSDART00000160465
|
opn6a
|
opsin 6, group member a |
| chr3_+_34220194 | 2.08 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr6_+_9867426 | 2.05 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr25_-_17552299 | 2.01 |
ENSDART00000154327
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr23_-_10137322 | 1.97 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr23_-_1587955 | 1.93 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr7_-_66864756 | 1.87 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr6_-_35401282 | 1.87 |
ENSDART00000127612
|
rgs5a
|
regulator of G protein signaling 5a |
| chr17_+_15845765 | 1.80 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr17_-_20979077 | 1.75 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr3_-_49504023 | 1.72 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr12_+_41991635 | 1.69 |
ENSDART00000186161
ENSDART00000192510 |
TCERG1L
|
transcription elongation regulator 1 like |
| chr20_+_29587995 | 1.67 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr25_+_192116 | 1.66 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr12_+_47044707 | 1.65 |
ENSDART00000186506
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr20_-_25436389 | 1.62 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr1_-_45177373 | 1.60 |
ENSDART00000143142
ENSDART00000034549 |
zgc:111983
|
zgc:111983 |
| chr22_+_38164486 | 1.57 |
ENSDART00000137521
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr20_-_18736281 | 1.51 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr17_-_33414781 | 1.49 |
ENSDART00000142203
ENSDART00000034638 |
ccdc28a
|
coiled-coil domain containing 28A |
| chr23_-_17509656 | 1.47 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr24_-_7777389 | 1.45 |
ENSDART00000138541
|
rpgrip1
|
RPGR interacting protein 1 |
| chr1_-_7894255 | 1.45 |
ENSDART00000167126
ENSDART00000145460 |
radil
|
Ras association and DIL domains |
| chr20_-_14680897 | 1.43 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr3_+_26144765 | 1.43 |
ENSDART00000146267
ENSDART00000043932 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr2_+_28995776 | 1.43 |
ENSDART00000138733
|
cdh12a
|
cadherin 12, type 2a (N-cadherin 2) |
| chr24_-_24271629 | 1.40 |
ENSDART00000135060
|
rps6ka3b
|
ribosomal protein S6 kinase, polypeptide 3b |
| chr3_-_16227683 | 1.35 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr4_-_14926637 | 1.35 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr3_+_14157090 | 1.34 |
ENSDART00000156766
|
si:ch211-108d22.2
|
si:ch211-108d22.2 |
| chr11_-_28148033 | 1.32 |
ENSDART00000177182
|
lactbl1b
|
lactamase, beta-like 1b |
| chr6_-_59381391 | 1.30 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr8_+_4337312 | 1.30 |
ENSDART00000182228
|
myl2b
|
myosin, light chain 2b, regulatory, cardiac, slow |
| chr7_-_57933736 | 1.30 |
ENSDART00000142580
|
ank2b
|
ankyrin 2b, neuronal |
| chr17_-_15657029 | 1.30 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr22_-_910926 | 1.29 |
ENSDART00000180075
|
FP016205.1
|
|
| chr24_-_38079261 | 1.28 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr15_+_15375607 | 1.27 |
ENSDART00000129751
|
si:ch211-105f12.2
|
si:ch211-105f12.2 |
| chr10_+_22134606 | 1.26 |
ENSDART00000155228
|
si:dkey-4c2.11
|
si:dkey-4c2.11 |
| chr2_-_1569250 | 1.25 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr7_-_9674073 | 1.22 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr18_-_36135799 | 1.21 |
ENSDART00000059344
|
b3gat1a
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) a |
| chr10_+_6013076 | 1.18 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr21_-_15046065 | 1.18 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr1_+_55758257 | 1.13 |
ENSDART00000139312
|
tecrb
|
trans-2,3-enoyl-CoA reductase b |
| chr18_-_1228688 | 1.13 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr11_+_36180349 | 1.13 |
ENSDART00000012940
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr7_-_35516251 | 1.12 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr2_+_30721070 | 1.08 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr22_+_38581012 | 1.06 |
ENSDART00000169239
|
BX088728.1
|
|
| chr24_-_24060632 | 1.06 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr20_-_9963713 | 1.04 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr21_-_2348838 | 1.03 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr19_+_24068223 | 1.00 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr25_+_22274642 | 1.00 |
ENSDART00000127099
|
nr2e3
|
nuclear receptor subfamily 2, group E, member 3 |
| chr9_-_29544720 | 1.00 |
ENSDART00000130317
|
arhgap20
|
Rho GTPase activating protein 20 |
| chr18_-_43884044 | 1.00 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr10_-_36738619 | 0.99 |
ENSDART00000093000
ENSDART00000157179 |
PLEKHB1
|
si:ch211-176g13.7 |
| chr2_-_52365251 | 0.98 |
ENSDART00000097716
|
tle2c
|
transducin like enhancer of split 2c |
| chr2_+_30878864 | 0.98 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr19_-_11846958 | 0.96 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr17_-_21368775 | 0.96 |
ENSDART00000058027
|
shtn1
|
shootin 1 |
| chr21_-_39327223 | 0.95 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr4_-_7212875 | 0.94 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr15_-_38154616 | 0.91 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr16_-_22585289 | 0.87 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr12_+_7865470 | 0.86 |
ENSDART00000161683
|
BX548028.1
|
|
| chr18_-_7948188 | 0.85 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr5_+_24287927 | 0.85 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr3_-_33574576 | 0.77 |
ENSDART00000184881
|
CR847537.1
|
|
| chr7_+_4384863 | 0.76 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr7_-_35515931 | 0.75 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr3_-_16227490 | 0.73 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr21_-_20733615 | 0.72 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr1_-_7893808 | 0.70 |
ENSDART00000110154
|
radil
|
Ras association and DIL domains |
| chr19_+_4061699 | 0.69 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr16_+_29555801 | 0.68 |
ENSDART00000169425
|
ensab
|
endosulfine alpha b |
| chr19_-_32944050 | 0.67 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr4_-_760560 | 0.67 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr3_-_30941362 | 0.60 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chrM_+_12897 | 0.59 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr6_-_19270484 | 0.59 |
ENSDART00000186894
ENSDART00000188709 |
zgc:174863
|
zgc:174863 |
| chr13_-_36525982 | 0.58 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr14_+_44545092 | 0.56 |
ENSDART00000175454
|
lingo2a
|
leucine rich repeat and Ig domain containing 2a |
| chr8_+_2656231 | 0.55 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr7_+_17063761 | 0.55 |
ENSDART00000182880
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr10_+_13279079 | 0.54 |
ENSDART00000135082
|
tmem267
|
transmembrane protein 267 |
| chr20_+_37295006 | 0.50 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr7_-_12821277 | 0.50 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr25_+_19041329 | 0.45 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr2_-_16986894 | 0.44 |
ENSDART00000145720
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr17_-_14559576 | 0.42 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr11_+_27973055 | 0.42 |
ENSDART00000135135
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr9_-_14084743 | 0.41 |
ENSDART00000056105
|
fer1l6
|
fer-1-like family member 6 |
| chr2_-_9818640 | 0.38 |
ENSDART00000139499
ENSDART00000165548 ENSDART00000012442 ENSDART00000046587 |
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr2_-_42071558 | 0.37 |
ENSDART00000142792
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr2_+_30531726 | 0.35 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr19_+_24882845 | 0.35 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr21_-_36948 | 0.34 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr3_+_52475058 | 0.32 |
ENSDART00000035867
|
si:ch211-241f5.3
|
si:ch211-241f5.3 |
| chr7_+_30970045 | 0.31 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr24_+_3478871 | 0.30 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr1_+_16573982 | 0.28 |
ENSDART00000166317
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr19_+_2275019 | 0.27 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr5_-_33274645 | 0.27 |
ENSDART00000188584
|
kyat1
|
kynurenine aminotransferase 1 |
| chr17_-_50010121 | 0.26 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr3_-_10970502 | 0.25 |
ENSDART00000127500
|
CR382337.1
|
|
| chr20_+_13533544 | 0.23 |
ENSDART00000143115
|
sytl3
|
synaptotagmin-like 3 |
| chr5_-_57526807 | 0.21 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr16_-_31675669 | 0.20 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr16_+_11297703 | 0.19 |
ENSDART00000125158
|
znf574
|
zinc finger protein 574 |
| chr22_-_20695237 | 0.17 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr24_-_1303553 | 0.17 |
ENSDART00000190984
|
nrp1a
|
neuropilin 1a |
| chr7_+_20031202 | 0.16 |
ENSDART00000052904
|
slc16a13
|
solute carrier family 16, member 13 (monocarboxylic acid transporter 13) |
| chr8_+_48848200 | 0.16 |
ENSDART00000130673
|
tp73
|
tumor protein p73 |
| chr5_+_67371650 | 0.13 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr8_+_21280360 | 0.13 |
ENSDART00000144488
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr5_+_8767290 | 0.12 |
ENSDART00000189377
|
CR854981.1
|
|
| chr23_-_28294763 | 0.11 |
ENSDART00000139537
|
znf385a
|
zinc finger protein 385A |
| chr15_-_14486534 | 0.10 |
ENSDART00000179368
|
numbl
|
numb homolog (Drosophila)-like |
| chr21_-_35419486 | 0.10 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr3_-_58798815 | 0.09 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr25_-_22889519 | 0.09 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr7_-_50517023 | 0.09 |
ENSDART00000073910
|
adamtsl5
|
ADAMTS like 5 |
| chr7_-_18656069 | 0.08 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr11_-_3366782 | 0.08 |
ENSDART00000127982
|
suox
|
sulfite oxidase |
| chr20_+_32473584 | 0.08 |
ENSDART00000192449
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr22_+_661505 | 0.06 |
ENSDART00000149460
|
elf3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr4_+_33012407 | 0.06 |
ENSDART00000151873
|
si:dkey-26h11.2
|
si:dkey-26h11.2 |
| chr24_+_5826005 | 0.04 |
ENSDART00000154930
|
si:ch211-157j23.5
|
si:ch211-157j23.5 |
| chr17_-_32863250 | 0.04 |
ENSDART00000167292
|
prox1a
|
prospero homeobox 1a |
| chr1_+_27690 | 0.03 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr7_-_17780048 | 0.03 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr20_+_36629173 | 0.02 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr18_-_25646286 | 0.02 |
ENSDART00000099511
ENSDART00000186890 |
si:ch211-13k12.2
|
si:ch211-13k12.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gsc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 1.9 | 22.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 1.1 | 6.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.9 | 6.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 2.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.4 | 1.2 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.4 | 1.4 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.3 | 1.7 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 1.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 7.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.3 | 13.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.3 | 1.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.7 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 1.0 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 2.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 2.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 8.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.2 | 1.0 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.2 | 2.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 1.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.2 | 5.1 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.2 | 1.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.6 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 2.1 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 3.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.7 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 2.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.8 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 2.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.7 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 2.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 2.0 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 2.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 3.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 1.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.6 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 1.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 10.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.6 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.1 | 3.8 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 1.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.2 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 3.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 2.6 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.1 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 2.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.6 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.7 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.3 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.4 | GO:0001503 | ossification(GO:0001503) |
| 0.0 | 5.1 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.4 | GO:0006821 | chloride transport(GO:0006821) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.9 | 13.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.6 | 9.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.5 | 2.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.5 | 18.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.4 | 3.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.4 | 1.4 | GO:0031673 | H zone(GO:0031673) |
| 0.3 | 4.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 3.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 6.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.0 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 8.5 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 2.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.8 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 3.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 2.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 3.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 2.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.6 | 2.8 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.5 | 6.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.5 | 9.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.5 | 1.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 1.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.3 | 1.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.3 | 1.3 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.3 | 1.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.3 | 13.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.3 | 1.4 | GO:0070004 | dipeptidase activity(GO:0016805) cysteine-type exopeptidase activity(GO:0070004) |
| 0.3 | 11.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 5.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 2.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.2 | 1.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 3.5 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 2.4 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.2 | 1.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 2.0 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 1.0 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 1.7 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 2.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 2.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 22.7 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.1 | 2.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 7.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 3.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.3 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.1 | 2.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 11.8 | GO:0015293 | symporter activity(GO:0015293) |
| 0.1 | 2.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.4 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 2.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 4.2 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 1.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 3.8 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 3.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.0 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 1.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 26.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 2.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 23.9 | REACTOME OPSINS | Genes involved in Opsins |
| 2.4 | 9.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 1.0 | 2.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.3 | 2.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 6.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.0 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |