Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
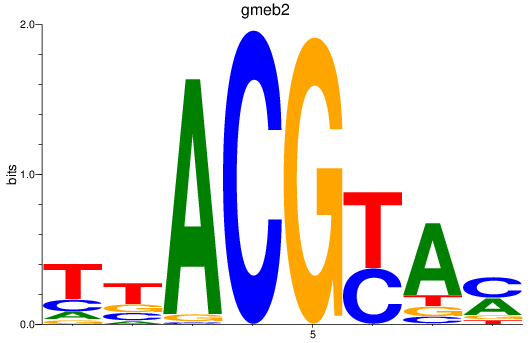
Results for gmeb2
Z-value: 0.61
Transcription factors associated with gmeb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb2
|
ENSDARG00000093240 | si_ch73-302a13.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GMEB2 | dr11_v1_chr23_-_41824460_41824460 | -0.72 | 5.1e-04 | Click! |
Activity profile of gmeb2 motif
Sorted Z-values of gmeb2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_20195350 | 3.28 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr5_-_67365333 | 1.62 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr14_+_10625112 | 1.60 |
ENSDART00000143377
ENSDART00000136480 |
nup62l
|
nucleoporin 62 like |
| chr2_-_17114852 | 1.49 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr21_+_6397463 | 1.45 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr5_+_26138313 | 1.41 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr3_-_49382896 | 1.36 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr10_-_17484762 | 1.20 |
ENSDART00000137905
ENSDART00000007961 |
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr16_+_40131473 | 1.12 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr11_-_36475124 | 1.12 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr12_-_43685802 | 1.07 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr2_+_56012016 | 0.94 |
ENSDART00000146160
ENSDART00000188702 |
loxl5b
|
lysyl oxidase-like 5b |
| chr17_-_31611692 | 0.93 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr6_-_52675630 | 0.92 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr2_-_17115256 | 0.92 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr25_+_21098990 | 0.91 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr16_+_14201401 | 0.90 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr10_-_14545658 | 0.90 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr7_-_39751540 | 0.86 |
ENSDART00000016803
|
grpel1
|
GrpE-like 1, mitochondrial |
| chr8_+_13064750 | 0.85 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr12_+_45238292 | 0.85 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr2_-_42128714 | 0.84 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr2_-_21170517 | 0.82 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr20_-_39367895 | 0.81 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr5_-_16139600 | 0.80 |
ENSDART00000051644
|
coq5
|
coenzyme Q5, methyltransferase |
| chr8_+_10304981 | 0.79 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr6_-_40455109 | 0.77 |
ENSDART00000103878
|
vhl
|
von Hippel-Lindau tumor suppressor |
| chr7_+_41313568 | 0.76 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr22_-_26353916 | 0.76 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr1_-_23274038 | 0.75 |
ENSDART00000181658
|
rfc1
|
replication factor C (activator 1) 1 |
| chr9_+_19529951 | 0.75 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr3_+_22335030 | 0.75 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr25_-_21066136 | 0.74 |
ENSDART00000109520
|
fbxl14a
|
F-box and leucine-rich repeat protein 14a |
| chr2_+_10821127 | 0.74 |
ENSDART00000145770
ENSDART00000174629 ENSDART00000081094 |
glmna
|
glomulin, FKBP associated protein a |
| chr25_+_21098675 | 0.74 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr7_-_7493758 | 0.73 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr6_-_39275793 | 0.73 |
ENSDART00000180477
ENSDART00000148531 |
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr1_-_23274393 | 0.72 |
ENSDART00000147800
ENSDART00000130277 ENSDART00000054340 ENSDART00000054338 |
rpl9
|
ribosomal protein L9 |
| chr22_-_621888 | 0.72 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr4_+_23126558 | 0.71 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr2_+_2168547 | 0.71 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr4_-_12914163 | 0.70 |
ENSDART00000140002
ENSDART00000145917 ENSDART00000141355 ENSDART00000067135 |
msrb3
|
methionine sulfoxide reductase B3 |
| chr24_+_39638555 | 0.69 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr2_+_10821579 | 0.67 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr16_+_23397785 | 0.66 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr8_-_4760723 | 0.66 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr1_+_9860381 | 0.66 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr1_+_51479128 | 0.66 |
ENSDART00000028018
|
meis1a
|
Meis homeobox 1 a |
| chr10_+_22034477 | 0.65 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr2_-_20923864 | 0.65 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr19_+_40026041 | 0.65 |
ENSDART00000017359
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr7_+_51795667 | 0.64 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr5_-_67365006 | 0.64 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr11_-_22372072 | 0.62 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr16_-_47427016 | 0.62 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr1_-_26063188 | 0.62 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr12_+_28956374 | 0.62 |
ENSDART00000033878
|
znf668
|
zinc finger protein 668 |
| chr4_-_11810365 | 0.61 |
ENSDART00000150529
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr17_-_51818659 | 0.61 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr6_-_39903393 | 0.60 |
ENSDART00000085945
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr23_-_33558161 | 0.58 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr7_+_66634167 | 0.57 |
ENSDART00000027616
|
eif4g2a
|
eukaryotic translation initiation factor 4, gamma 2a |
| chr16_+_41015163 | 0.57 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr16_+_31921812 | 0.56 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr13_+_42309688 | 0.56 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr6_-_10728057 | 0.56 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr2_+_32826235 | 0.55 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr1_-_31516091 | 0.55 |
ENSDART00000139828
ENSDART00000146567 |
cenpk
|
centromere protein K |
| chr17_-_49407091 | 0.53 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr13_+_3938526 | 0.53 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr9_-_23242684 | 0.53 |
ENSDART00000053282
ENSDART00000179770 |
ccnt2a
|
cyclin T2a |
| chr11_-_14813029 | 0.52 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr8_+_10305400 | 0.52 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_+_38055529 | 0.52 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr4_-_11810799 | 0.52 |
ENSDART00000014153
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr9_+_21819082 | 0.51 |
ENSDART00000136902
ENSDART00000101991 |
txndc9
|
thioredoxin domain containing 9 |
| chr12_+_33460794 | 0.51 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr1_-_26062798 | 0.51 |
ENSDART00000185348
|
pdcd4a
|
programmed cell death 4a |
| chr15_-_23692359 | 0.50 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr6_-_37745508 | 0.50 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr23_+_20640484 | 0.50 |
ENSDART00000054691
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr5_-_54395488 | 0.48 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr23_-_33738570 | 0.48 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr23_-_33738945 | 0.46 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr18_-_14879135 | 0.46 |
ENSDART00000099701
|
selenoo1
|
selenoprotein O1 |
| chr9_+_25840720 | 0.46 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr8_-_30944465 | 0.45 |
ENSDART00000128792
ENSDART00000191717 ENSDART00000049944 |
smarcb1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1a |
| chr1_+_23274710 | 0.45 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr10_+_28428222 | 0.45 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr22_-_817479 | 0.44 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr7_+_55292959 | 0.44 |
ENSDART00000147539
ENSDART00000073555 |
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr13_+_30035253 | 0.44 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr8_+_20455134 | 0.43 |
ENSDART00000079618
|
rexo1
|
REX1, RNA exonuclease 1 homolog |
| chr14_-_21932403 | 0.43 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr20_-_37933237 | 0.42 |
ENSDART00000142567
ENSDART00000036371 ENSDART00000061445 |
angel2
|
angel homolog 2 (Drosophila) |
| chr22_-_22301929 | 0.42 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr13_-_36535128 | 0.42 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr7_-_23894879 | 0.42 |
ENSDART00000123203
|
vbp1
|
von Hippel-Lindau binding protein 1 |
| chr12_+_638435 | 0.42 |
ENSDART00000152508
|
si:ch211-176g6.1
|
si:ch211-176g6.1 |
| chr5_-_23596339 | 0.41 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr18_+_49248389 | 0.40 |
ENSDART00000059285
ENSDART00000142004 ENSDART00000132751 |
yif1b
|
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr22_+_20145036 | 0.40 |
ENSDART00000137624
|
eef2a.2
|
eukaryotic translation elongation factor 2a, tandem duplicate 2 |
| chr21_-_44772738 | 0.39 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr13_+_36923052 | 0.39 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr20_-_36809059 | 0.39 |
ENSDART00000062925
|
slc25a27
|
solute carrier family 25, member 27 |
| chr15_-_31406093 | 0.38 |
ENSDART00000123444
|
or111-8
|
odorant receptor, family D, subfamily 111, member 8 |
| chr17_+_37932433 | 0.38 |
ENSDART00000185349
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr25_-_8513255 | 0.38 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr5_-_12063381 | 0.38 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr13_+_21676235 | 0.38 |
ENSDART00000137804
ENSDART00000134950 ENSDART00000129653 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr1_+_49415281 | 0.37 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_-_12486076 | 0.37 |
ENSDART00000146195
|
lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr4_+_9011448 | 0.37 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr9_-_23747264 | 0.36 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr12_+_17603528 | 0.36 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr2_+_52049239 | 0.35 |
ENSDART00000036813
|
ccdc94
|
coiled-coil domain containing 94 |
| chr3_+_26342768 | 0.35 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr5_+_25585869 | 0.35 |
ENSDART00000138060
|
si:dkey-229d2.7
|
si:dkey-229d2.7 |
| chr24_-_7826489 | 0.34 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr3_+_53156813 | 0.34 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr10_+_20070178 | 0.34 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr20_-_9199721 | 0.34 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr3_+_46764022 | 0.34 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr15_+_32798333 | 0.33 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr7_+_52761841 | 0.32 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr25_-_28668776 | 0.32 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr15_-_43284021 | 0.32 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr2_-_32486080 | 0.32 |
ENSDART00000110821
|
ttc19
|
tetratricopeptide repeat domain 19 |
| chr17_-_48805159 | 0.31 |
ENSDART00000103662
|
kif6
|
kinesin family member 6 |
| chr17_-_6451801 | 0.31 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr18_-_40481028 | 0.30 |
ENSDART00000134177
|
zgc:101040
|
zgc:101040 |
| chr22_-_621609 | 0.30 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr8_-_4618653 | 0.29 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr3_+_59880317 | 0.29 |
ENSDART00000166922
ENSDART00000108647 |
alyref
|
Aly/REF export factor |
| chr17_-_36860988 | 0.29 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr8_+_53311965 | 0.29 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr20_-_32405440 | 0.28 |
ENSDART00000062978
ENSDART00000153411 |
afg1lb
|
AFG1 like ATPase b |
| chr15_+_5116179 | 0.28 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr20_-_31743817 | 0.28 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr3_+_46763745 | 0.28 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_25814097 | 0.27 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr18_-_20674121 | 0.27 |
ENSDART00000005145
|
asz1
|
ankyrin repeat, SAM and basic leucine zipper domain containing 1 |
| chr9_+_23049500 | 0.26 |
ENSDART00000012478
|
mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr23_+_17522867 | 0.26 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr18_+_30847237 | 0.25 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr12_+_11650146 | 0.25 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr12_-_27588299 | 0.25 |
ENSDART00000178023
ENSDART00000066282 |
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr17_-_12058171 | 0.25 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr20_+_38837238 | 0.24 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr16_-_47426482 | 0.24 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr6_-_51541488 | 0.23 |
ENSDART00000156336
|
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr8_-_32354677 | 0.23 |
ENSDART00000138268
ENSDART00000133245 ENSDART00000179677 ENSDART00000174450 |
ipo11
|
importin 11 |
| chr16_-_20294236 | 0.23 |
ENSDART00000059623
|
plekha8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr1_+_26105141 | 0.21 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr17_+_12058509 | 0.21 |
ENSDART00000150209
|
tfb2m
|
transcription factor B2, mitochondrial |
| chr8_+_1239 | 0.21 |
ENSDART00000067666
ENSDART00000192564 ENSDART00000192612 ENSDART00000190683 |
tmed7
|
transmembrane p24 trafficking protein 7 |
| chr16_-_29557338 | 0.21 |
ENSDART00000058888
|
hormad1
|
HORMA domain containing 1 |
| chr14_-_30808174 | 0.20 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr22_-_22301672 | 0.19 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr15_-_31364729 | 0.19 |
ENSDART00000185386
|
or111-2
|
odorant receptor, family D, subfamily 111, member 2 |
| chr4_+_8638622 | 0.19 |
ENSDART00000186829
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr25_-_6389713 | 0.19 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr1_-_49521407 | 0.19 |
ENSDART00000189845
ENSDART00000143474 |
zp3c
|
zona pellucida glycoprotein 3c |
| chr4_+_9011825 | 0.19 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr11_+_24298432 | 0.19 |
ENSDART00000138487
|
si:dkey-76p14.2
|
si:dkey-76p14.2 |
| chr17_-_25831569 | 0.18 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr12_-_10409961 | 0.17 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr7_-_55539738 | 0.17 |
ENSDART00000168721
ENSDART00000013796 ENSDART00000148514 |
aprt
|
adenine phosphoribosyltransferase |
| chr8_-_19467011 | 0.17 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr22_+_24214665 | 0.17 |
ENSDART00000163980
ENSDART00000167996 |
glrx2
|
glutaredoxin 2 |
| chr11_+_43419809 | 0.17 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr14_+_8947282 | 0.16 |
ENSDART00000047993
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr14_-_43000836 | 0.16 |
ENSDART00000162714
|
pcdh10b
|
protocadherin 10b |
| chr3_+_46764278 | 0.16 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr11_-_44945636 | 0.15 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr12_+_3571770 | 0.14 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr16_-_17300030 | 0.14 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr7_-_38570878 | 0.14 |
ENSDART00000139187
ENSDART00000134570 ENSDART00000041055 |
celf1
|
cugbp, Elav-like family member 1 |
| chr20_-_20410029 | 0.13 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr13_-_21688176 | 0.13 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr15_+_11683114 | 0.13 |
ENSDART00000168233
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr6_+_46668717 | 0.12 |
ENSDART00000064853
|
oxtrl
|
oxytocin receptor like |
| chr5_+_24245682 | 0.12 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr6_-_1223551 | 0.11 |
ENSDART00000182112
|
CABZ01063034.1
|
|
| chr24_+_37484661 | 0.11 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr6_-_25201810 | 0.11 |
ENSDART00000168683
|
lrrc8c
|
leucine rich repeat containing 8 VRAC subunit C |
| chr2_+_26647472 | 0.10 |
ENSDART00000145415
ENSDART00000157409 |
ttpa
|
tocopherol (alpha) transfer protein |
| chr12_-_11649690 | 0.10 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr11_-_41132296 | 0.10 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr8_+_30112655 | 0.10 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr24_+_34069675 | 0.09 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr18_+_5917625 | 0.09 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr3_-_40681001 | 0.09 |
ENSDART00000154929
|
si:dkey-33c3.1
|
si:dkey-33c3.1 |
| chr1_+_27024068 | 0.09 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr14_-_22495604 | 0.08 |
ENSDART00000137167
|
si:ch211-107m4.1
|
si:ch211-107m4.1 |
| chr9_+_24920677 | 0.08 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_-_10769097 | 0.07 |
ENSDART00000189967
|
tmbim1b
|
transmembrane BAX inhibitor motif containing 1b |
| chr25_+_32473277 | 0.07 |
ENSDART00000146451
|
sqor
|
sulfide quinone oxidoreductase |
| chr21_+_15704556 | 0.06 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr25_-_17494364 | 0.06 |
ENSDART00000154134
|
lrrc29
|
leucine rich repeat containing 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.5 | 1.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.5 | 2.3 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.4 | 1.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 | 0.8 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.2 | 2.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 0.9 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.2 | 0.6 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 0.6 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 0.5 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.2 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 0.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.7 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.8 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 2.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.7 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.7 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.1 | 0.5 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.2 | GO:0006168 | adenine salvage(GO:0006168) |
| 0.0 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.8 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.2 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 1.9 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 | 0.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.5 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.0 | 0.4 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.4 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.4 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 1.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.8 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 0.5 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.2 | 1.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.5 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.7 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.6 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.5 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.9 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 1.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 1.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.8 | 2.3 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.5 | 2.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.5 | 1.4 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.8 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 1.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.3 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0034246 | core DNA-dependent RNA polymerase binding promoter specificity activity(GO:0000996) mitochondrial RNA polymerase binding promoter specificity activity(GO:0034246) |
| 0.1 | 0.9 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.4 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 1.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.8 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 2.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.9 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |