Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
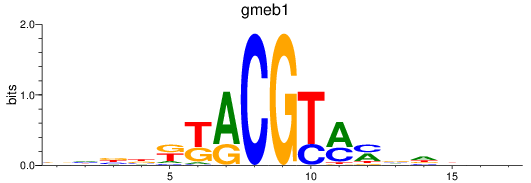
Results for gmeb1
Z-value: 0.46
Transcription factors associated with gmeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb1
|
ENSDARG00000062060 | glucocorticoid modulatory element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gmeb1 | dr11_v1_chr19_+_16064439_16064439 | 0.80 | 3.8e-05 | Click! |
Activity profile of gmeb1 motif
Sorted Z-values of gmeb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_45238292 | 1.15 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr25_+_34845115 | 1.13 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr22_+_22437561 | 1.01 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr3_-_15999501 | 0.98 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr21_+_6397463 | 0.87 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr3_-_16000168 | 0.87 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr16_-_41535690 | 0.73 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr4_+_23126558 | 0.72 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr8_+_10305400 | 0.64 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr10_-_20523405 | 0.59 |
ENSDART00000114824
|
ddhd2
|
DDHD domain containing 2 |
| chr8_+_19489854 | 0.57 |
ENSDART00000184671
ENSDART00000011258 |
npl
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| chr25_-_34845302 | 0.56 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr16_+_23397785 | 0.49 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr22_-_621888 | 0.48 |
ENSDART00000135829
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr9_-_306515 | 0.44 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr5_+_30588856 | 0.40 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr1_+_19515228 | 0.38 |
ENSDART00000103091
|
clrn2
|
clarin 2 |
| chr25_+_22281441 | 0.35 |
ENSDART00000089516
|
stoml1
|
stomatin (EPB72)-like 1 |
| chr23_-_9807546 | 0.35 |
ENSDART00000136740
ENSDART00000004474 |
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr14_-_21932403 | 0.35 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr12_+_11650146 | 0.35 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr5_+_6955900 | 0.32 |
ENSDART00000099417
|
CABZ01041962.1
|
|
| chr14_-_31489410 | 0.26 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr12_-_2869565 | 0.24 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr12_+_3571770 | 0.24 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr22_-_621609 | 0.22 |
ENSDART00000137264
ENSDART00000106636 |
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr20_+_27427112 | 0.19 |
ENSDART00000153459
|
si:dkey-22l1.1
|
si:dkey-22l1.1 |
| chr10_+_39091716 | 0.18 |
ENSDART00000193072
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr3_+_46764278 | 0.17 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr21_-_8153165 | 0.16 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr25_+_18711804 | 0.15 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr12_-_11649690 | 0.15 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr2_-_3045861 | 0.15 |
ENSDART00000105818
ENSDART00000187575 |
guk1a
|
guanylate kinase 1a |
| chr16_-_24815091 | 0.15 |
ENSDART00000154269
ENSDART00000131025 |
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr17_-_25831569 | 0.13 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr10_+_26990095 | 0.13 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 1.9 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.2 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.3 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.7 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.6 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |