Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
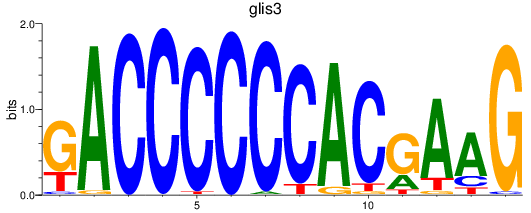
Results for glis3
Z-value: 0.22
Transcription factors associated with glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis3
|
ENSDARG00000069726 | GLIS family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| glis3 | dr11_v1_chr10_-_690072_690170 | -0.84 | 6.5e-06 | Click! |
Activity profile of glis3 motif
Sorted Z-values of glis3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_30704075 | 1.22 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr8_+_46641314 | 0.98 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr5_+_67662430 | 0.41 |
ENSDART00000137700
ENSDART00000142586 |
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr20_-_26588736 | 0.36 |
ENSDART00000134337
|
exoc2
|
exocyst complex component 2 |
| chr8_-_15182232 | 0.34 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr5_+_25760112 | 0.32 |
ENSDART00000088011
ENSDART00000182046 |
tmem2
|
transmembrane protein 2 |
| chr15_-_41245962 | 0.31 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr3_-_25106878 | 0.31 |
ENSDART00000142503
|
rbx1
|
ring-box 1, E3 ubiquitin protein ligase |
| chr4_+_77981553 | 0.28 |
ENSDART00000174108
ENSDART00000122459 |
terfa
|
telomeric repeat binding factor a |
| chr17_+_8925232 | 0.24 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr3_+_30190419 | 0.24 |
ENSDART00000157320
|
akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr21_+_20950161 | 0.18 |
ENSDART00000079698
|
htr1ab
|
5-hydroxytryptamine (serotonin) receptor 1A b |
| chr5_+_45000597 | 0.13 |
ENSDART00000051146
|
dmrt3a
|
doublesex and mab-3 related transcription factor 3a |
| chr3_-_53092509 | 0.10 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr18_+_33100606 | 0.09 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr21_+_20949976 | 0.09 |
ENSDART00000135342
|
htr1ab
|
5-hydroxytryptamine (serotonin) receptor 1A b |
| chr14_-_34044369 | 0.04 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr17_+_49500820 | 0.02 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr18_-_41375120 | 0.02 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr10_+_29260096 | 0.02 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of glis3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.1 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.0 | 0.4 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |