Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
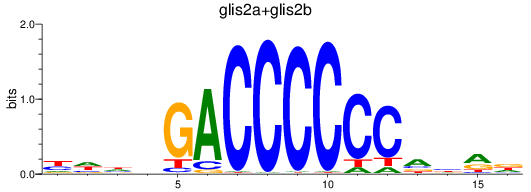
Results for glis2a+glis2b
Z-value: 0.57
Transcription factors associated with glis2a+glis2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis2a
|
ENSDARG00000078388 | GLIS family zinc finger 2a |
|
glis2b
|
ENSDARG00000100232 | GLIS family zinc finger 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| glis2b | dr11_v1_chr3_+_12334509_12334509 | 0.88 | 8.2e-07 | Click! |
| glis2a | dr11_v1_chr22_-_26558166_26558166 | 0.77 | 1.3e-04 | Click! |
Activity profile of glis2a+glis2b motif
Sorted Z-values of glis2a+glis2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_51079504 | 1.60 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr20_-_25518488 | 1.22 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr13_-_638485 | 1.11 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr2_+_51783120 | 1.07 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr24_-_38574631 | 0.84 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr25_-_997894 | 0.76 |
ENSDART00000169011
|
znf609a
|
zinc finger protein 609a |
| chr3_+_34220194 | 0.74 |
ENSDART00000145859
|
slc25a23b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23b |
| chr16_+_3004422 | 0.69 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr7_+_57725708 | 0.68 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr24_+_37723362 | 0.67 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr6_-_7769178 | 0.66 |
ENSDART00000191701
ENSDART00000149823 |
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr11_+_27968834 | 0.61 |
ENSDART00000147984
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr19_-_48336535 | 0.61 |
ENSDART00000162752
|
si:ch73-359m17.6
|
si:ch73-359m17.6 |
| chr21_+_25643880 | 0.59 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr24_-_35707552 | 0.57 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr20_-_1239596 | 0.55 |
ENSDART00000049063
ENSDART00000140650 |
ankrd6b
|
ankyrin repeat domain 6b |
| chr7_+_38509333 | 0.53 |
ENSDART00000153482
|
slc7a9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr16_-_26435431 | 0.50 |
ENSDART00000187526
|
megf8
|
multiple EGF-like-domains 8 |
| chr17_-_4245311 | 0.49 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr5_+_9405283 | 0.49 |
ENSDART00000127306
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr23_-_36934944 | 0.47 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr22_-_18116635 | 0.44 |
ENSDART00000005724
|
ncanb
|
neurocan b |
| chr19_-_36640677 | 0.43 |
ENSDART00000189743
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr17_+_23578316 | 0.38 |
ENSDART00000149281
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr9_+_20781047 | 0.37 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr23_-_29004753 | 0.36 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr4_-_17725008 | 0.36 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr17_-_4245902 | 0.34 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr17_-_3291369 | 0.33 |
ENSDART00000181840
|
CABZ01007222.1
|
|
| chr12_+_2676878 | 0.31 |
ENSDART00000185909
|
antxr1c
|
anthrax toxin receptor 1c |
| chr23_-_29003864 | 0.31 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr5_-_1047222 | 0.30 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr6_+_38845848 | 0.29 |
ENSDART00000184907
|
stk35l
|
serine/threonine kinase 35, like |
| chr13_+_39208542 | 0.27 |
ENSDART00000147971
|
fam135a
|
family with sequence similarity 135, member A |
| chr24_+_25471196 | 0.27 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr23_+_39481537 | 0.27 |
ENSDART00000187222
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr2_+_13722116 | 0.26 |
ENSDART00000155015
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr5_-_22001003 | 0.22 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr7_+_11390462 | 0.22 |
ENSDART00000114383
|
tlnrd1
|
talin rod domain containing 1 |
| chr16_-_45854882 | 0.21 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr15_-_97205 | 0.20 |
ENSDART00000158734
|
BACE1
|
beta-secretase 1 |
| chr19_+_20540843 | 0.19 |
ENSDART00000192500
|
kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr21_-_588858 | 0.18 |
ENSDART00000168983
|
TMEM38B
|
transmembrane protein 38B |
| chr25_+_16912243 | 0.18 |
ENSDART00000186579
|
zgc:77158
|
zgc:77158 |
| chr18_+_17151916 | 0.16 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr2_+_13721888 | 0.13 |
ENSDART00000089618
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr10_-_13116337 | 0.13 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr23_-_45705525 | 0.12 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr17_-_44968177 | 0.12 |
ENSDART00000075510
|
ngb
|
neuroglobin |
| chr15_+_518092 | 0.12 |
ENSDART00000154594
|
pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr13_-_24880525 | 0.11 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr20_+_38285671 | 0.09 |
ENSDART00000061432
|
ccl38a.4
|
chemokine (C-C motif) ligand 38, duplicate 4 |
| chr3_+_40409100 | 0.08 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr7_-_19642417 | 0.08 |
ENSDART00000160936
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr10_-_1961576 | 0.06 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr7_-_24838857 | 0.05 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr5_+_38886499 | 0.05 |
ENSDART00000076845
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr6_+_38845697 | 0.04 |
ENSDART00000053187
|
stk35l
|
serine/threonine kinase 35, like |
| chr17_-_20717845 | 0.03 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr4_-_59152337 | 0.03 |
ENSDART00000156770
|
znf1149
|
zinc finger protein 1149 |
| chr3_-_61592417 | 0.03 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr3_-_33113879 | 0.03 |
ENSDART00000044677
|
rarab
|
retinoic acid receptor, alpha b |
| chr23_+_42810055 | 0.01 |
ENSDART00000186647
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr23_+_43668756 | 0.00 |
ENSDART00000112598
|
otud4
|
OTU deubiquitinase 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of glis2a+glis2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.7 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.6 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 1.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.0 | 0.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0055038 | cleavage furrow(GO:0032154) recycling endosome membrane(GO:0055038) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.4 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.8 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.2 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 1.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |