Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
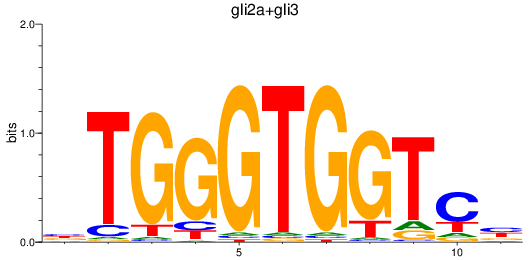
Results for gli2a+gli3
Z-value: 0.76
Transcription factors associated with gli2a+gli3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2a
|
ENSDARG00000025641 | GLI family zinc finger 2a |
|
gli3
|
ENSDARG00000052131 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli2a | dr11_v1_chr9_+_37141836_37141836 | -0.87 | 1.7e-06 | Click! |
| gli3 | dr11_v1_chr24_-_11821505_11821505 | -0.80 | 4.1e-05 | Click! |
Activity profile of gli2a+gli3 motif
Sorted Z-values of gli2a+gli3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_3252950 | 5.60 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr16_+_52512025 | 4.00 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr10_-_22854758 | 3.16 |
ENSDART00000079449
|
and3
|
actinodin3 |
| chr5_+_51594209 | 2.24 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr6_-_35446110 | 2.23 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr13_+_3954540 | 2.01 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr21_+_10739846 | 1.95 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr19_-_40192249 | 1.95 |
ENSDART00000051972
|
grn1
|
granulin 1 |
| chr2_+_51783120 | 1.94 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr23_-_27875140 | 1.85 |
ENSDART00000143662
|
ankrd33aa
|
ankyrin repeat domain 33Aa |
| chr17_-_51202339 | 1.75 |
ENSDART00000167117
|
si:ch1073-469d17.2
|
si:ch1073-469d17.2 |
| chr23_+_28582865 | 1.64 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr2_-_30784502 | 1.56 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr25_-_210730 | 1.56 |
ENSDART00000187580
|
FP236318.1
|
|
| chr12_-_464007 | 1.55 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr15_+_7992906 | 1.53 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr2_-_57707039 | 1.52 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr4_+_72797711 | 1.47 |
ENSDART00000190934
ENSDART00000163236 |
MYRFL
|
myelin regulatory factor-like |
| chr19_-_10243148 | 1.47 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr4_-_55785014 | 1.46 |
ENSDART00000181348
|
CT583728.24
|
|
| chr4_-_55801210 | 1.46 |
ENSDART00000186943
|
CT583728.13
|
|
| chr4_-_55829342 | 1.46 |
ENSDART00000185611
|
CT583728.26
|
|
| chr20_+_27194833 | 1.44 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr4_-_17741513 | 1.43 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr25_-_17403598 | 1.43 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr16_+_20220225 | 1.32 |
ENSDART00000182208
|
colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr23_+_20422661 | 1.28 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr6_-_10320676 | 1.26 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr23_+_43834376 | 1.25 |
ENSDART00000148989
|
si:ch211-149b19.4
|
si:ch211-149b19.4 |
| chr5_-_20678300 | 1.24 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr10_-_35542071 | 1.24 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr4_-_75175407 | 1.22 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr18_+_9641210 | 1.21 |
ENSDART00000182024
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr10_+_29963518 | 1.11 |
ENSDART00000011317
ENSDART00000099964 ENSDART00000182990 ENSDART00000113912 |
ntm
|
neurotrimin |
| chr1_-_19845378 | 1.09 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr15_+_16525126 | 1.08 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr19_+_9050852 | 1.06 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr20_-_915234 | 1.05 |
ENSDART00000164816
|
cnr1
|
cannabinoid receptor 1 |
| chr1_-_17797802 | 1.04 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr1_-_46862190 | 1.04 |
ENSDART00000145167
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr25_+_30298377 | 1.02 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr18_+_9635178 | 1.00 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr6_+_54358529 | 0.99 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr22_-_10487490 | 0.93 |
ENSDART00000064798
|
aspn
|
asporin (LRR class 1) |
| chr4_+_5132951 | 0.92 |
ENSDART00000103279
|
ccnd2b
|
cyclin D2, b |
| chr16_-_31644545 | 0.90 |
ENSDART00000181634
|
CR855311.5
|
|
| chr1_+_9557212 | 0.89 |
ENSDART00000111131
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr5_-_17876709 | 0.88 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr7_-_2129076 | 0.87 |
ENSDART00000182385
|
si:cabz01007807.1
|
si:cabz01007807.1 |
| chr11_+_30314885 | 0.87 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr12_+_24562667 | 0.86 |
ENSDART00000056256
|
nrxn1a
|
neurexin 1a |
| chr16_+_42667560 | 0.86 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr16_+_37470717 | 0.84 |
ENSDART00000112003
ENSDART00000188431 ENSDART00000192837 |
adgrb1a
|
adhesion G protein-coupled receptor B1a |
| chr2_+_50738325 | 0.80 |
ENSDART00000114182
|
fyco1b
|
FYVE and coiled-coil domain containing 1b |
| chr22_+_38173960 | 0.80 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr8_-_47800754 | 0.80 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr5_+_13521081 | 0.79 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr9_-_46072805 | 0.79 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr8_+_31119548 | 0.79 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr2_-_30784198 | 0.78 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr3_-_46410387 | 0.78 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr5_+_36768674 | 0.75 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr18_+_16963881 | 0.74 |
ENSDART00000147583
|
si:ch211-242e8.1
|
si:ch211-242e8.1 |
| chr7_+_73780936 | 0.73 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr25_-_28384954 | 0.71 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr4_-_2219705 | 0.68 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr5_-_58112032 | 0.68 |
ENSDART00000016418
|
drd2b
|
dopamine receptor D2b |
| chr17_+_30587333 | 0.65 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr21_-_30935262 | 0.63 |
ENSDART00000172390
|
stim1b
|
stromal interaction molecule 1b |
| chr17_-_7371564 | 0.62 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr8_-_40075983 | 0.62 |
ENSDART00000141455
|
ggt1a
|
gamma-glutamyltransferase 1a |
| chr19_+_32201991 | 0.62 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr8_+_47342586 | 0.61 |
ENSDART00000007624
|
plch2a
|
phospholipase C, eta 2a |
| chr14_+_25464681 | 0.61 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr6_+_10094061 | 0.60 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr16_-_4026265 | 0.59 |
ENSDART00000149339
|
si:ch211-175f12.2
|
si:ch211-175f12.2 |
| chr5_+_36439405 | 0.58 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr8_-_49201258 | 0.58 |
ENSDART00000114584
ENSDART00000142172 |
zgc:171501
|
zgc:171501 |
| chr20_-_38610360 | 0.58 |
ENSDART00000168783
ENSDART00000164638 |
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr2_+_54086436 | 0.57 |
ENSDART00000174581
|
CU179656.1
|
|
| chr10_-_44411032 | 0.56 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr10_+_2232023 | 0.55 |
ENSDART00000097695
|
cntnap3
|
contactin associated protein like 3 |
| chr7_-_15413019 | 0.55 |
ENSDART00000172147
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr8_-_18316482 | 0.53 |
ENSDART00000190156
|
rnf220b
|
ring finger protein 220b |
| chr16_+_50741154 | 0.53 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr12_-_46145635 | 0.50 |
ENSDART00000074682
|
zgc:153932
|
zgc:153932 |
| chr8_-_7078575 | 0.50 |
ENSDART00000137413
|
si:ch73-233m11.2
|
si:ch73-233m11.2 |
| chr25_+_7461624 | 0.50 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr1_+_7956030 | 0.50 |
ENSDART00000159655
|
CR855320.1
|
|
| chr17_+_31580960 | 0.49 |
ENSDART00000188157
|
si:dkey-13p1.4
|
si:dkey-13p1.4 |
| chr20_-_35052823 | 0.47 |
ENSDART00000153033
|
kif26bb
|
kinesin family member 26Bb |
| chr13_-_15799391 | 0.47 |
ENSDART00000124688
|
bag5
|
BCL2 associated athanogene 5 |
| chr21_+_22985078 | 0.46 |
ENSDART00000156491
|
lpar6b
|
lysophosphatidic acid receptor 6b |
| chr2_+_47718605 | 0.46 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr3_-_19091024 | 0.45 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr20_-_7000225 | 0.43 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr2_+_48638827 | 0.43 |
ENSDART00000163634
|
CR381673.2
|
|
| chr5_+_20437124 | 0.43 |
ENSDART00000159001
|
tmem119a
|
transmembrane protein 119a |
| chr14_+_46020282 | 0.42 |
ENSDART00000190087
|
c1qtnf2
|
C1q and TNF related 2 |
| chr21_-_5856050 | 0.42 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr3_+_5083407 | 0.41 |
ENSDART00000146883
|
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr12_-_26582607 | 0.41 |
ENSDART00000045186
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr7_+_73801377 | 0.40 |
ENSDART00000184051
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr19_+_56351 | 0.40 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr6_+_7123980 | 0.40 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr14_+_25465346 | 0.40 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr10_+_26747755 | 0.39 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr22_-_188102 | 0.39 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr3_-_18805225 | 0.38 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr7_-_15413188 | 0.37 |
ENSDART00000185980
|
si:ch211-106j24.1
|
si:ch211-106j24.1 |
| chr20_+_10498704 | 0.37 |
ENSDART00000192985
|
zgc:100997
|
zgc:100997 |
| chr24_-_39354829 | 0.36 |
ENSDART00000169108
|
kcnj12a
|
potassium inwardly-rectifying channel, subfamily J, member 12a |
| chr21_-_26520629 | 0.35 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr20_-_6131686 | 0.35 |
ENSDART00000145964
ENSDART00000086578 ENSDART00000164090 |
pum2
|
pumilio RNA-binding family member 2 |
| chr9_-_11655031 | 0.34 |
ENSDART00000044314
|
itgav
|
integrin, alpha V |
| chr17_-_34952562 | 0.33 |
ENSDART00000021128
|
kidins220a
|
kinase D-interacting substrate 220a |
| chr15_+_42431198 | 0.33 |
ENSDART00000189951
ENSDART00000089694 |
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr4_-_38476901 | 0.32 |
ENSDART00000167124
|
si:ch211-209n20.59
|
si:ch211-209n20.59 |
| chr17_-_8562586 | 0.32 |
ENSDART00000154257
ENSDART00000157308 |
fzd3b
|
frizzled class receptor 3b |
| chr25_+_32473433 | 0.30 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr10_-_24868581 | 0.29 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr18_+_22793743 | 0.29 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr15_-_19128705 | 0.27 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr13_-_45201300 | 0.27 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr19_-_26235219 | 0.27 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr23_-_41824460 | 0.26 |
ENSDART00000131235
|
GMEB2
|
si:ch73-302a13.2 |
| chr4_+_4902392 | 0.24 |
ENSDART00000133866
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr9_-_33136990 | 0.22 |
ENSDART00000131742
|
slc5a3b
|
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3b |
| chr11_-_23025949 | 0.22 |
ENSDART00000184859
ENSDART00000167818 ENSDART00000046122 ENSDART00000193120 |
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr14_-_41091667 | 0.22 |
ENSDART00000170042
|
drp2
|
dystrophin related protein 2 |
| chr24_-_6258869 | 0.22 |
ENSDART00000006636
ENSDART00000178667 |
myo3a
|
myosin IIIA |
| chr6_-_15639087 | 0.21 |
ENSDART00000128940
ENSDART00000183092 |
mlpha
|
melanophilin a |
| chr10_+_18952271 | 0.21 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr23_-_14057191 | 0.21 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
| chr25_+_22730490 | 0.21 |
ENSDART00000149455
|
abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr3_+_57991074 | 0.21 |
ENSDART00000076077
|
myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr7_+_34790768 | 0.18 |
ENSDART00000075116
|
si:dkey-148a17.6
|
si:dkey-148a17.6 |
| chr11_+_36355348 | 0.18 |
ENSDART00000145427
|
sypl2a
|
synaptophysin-like 2a |
| chr25_-_24247584 | 0.17 |
ENSDART00000046349
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr17_-_4252221 | 0.17 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr3_+_56574623 | 0.17 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr20_-_35052464 | 0.15 |
ENSDART00000037195
|
kif26bb
|
kinesin family member 26Bb |
| chr24_-_25184553 | 0.15 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr14_+_30774515 | 0.15 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr11_+_440305 | 0.15 |
ENSDART00000082517
|
rab43
|
RAB43, member RAS oncogene family |
| chr8_-_14484599 | 0.14 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr19_+_7954679 | 0.14 |
ENSDART00000150909
|
si:dkey-266f7.10
|
si:dkey-266f7.10 |
| chr21_-_38619305 | 0.13 |
ENSDART00000139178
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr18_-_17485419 | 0.12 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr7_-_8374950 | 0.12 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr10_+_40235959 | 0.12 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
| chr1_+_10129099 | 0.11 |
ENSDART00000187740
|
rbm46
|
RNA binding motif protein 46 |
| chr8_+_28467893 | 0.11 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr14_+_49152341 | 0.11 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr5_-_23715861 | 0.10 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr5_+_20599658 | 0.10 |
ENSDART00000143019
|
CMKLR1 (1 of many)
|
si:ch211-240b21.4 |
| chr3_-_51109286 | 0.09 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr17_+_6563307 | 0.09 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr16_-_7827517 | 0.09 |
ENSDART00000189975
|
tcaim
|
T cell activation inhibitor, mitochondrial |
| chr22_+_38762693 | 0.09 |
ENSDART00000015016
ENSDART00000150187 |
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr24_+_7828613 | 0.08 |
ENSDART00000135161
|
zgc:101569
|
zgc:101569 |
| chr20_+_2669017 | 0.08 |
ENSDART00000058777
|
slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr6_-_8480815 | 0.08 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr13_-_865193 | 0.07 |
ENSDART00000187053
|
AL929536.7
|
|
| chr24_-_10828560 | 0.05 |
ENSDART00000132282
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr5_-_31926906 | 0.05 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr14_-_215051 | 0.04 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr9_-_12736089 | 0.04 |
ENSDART00000088042
|
myo10l3
|
myosin X-like 3 |
| chr14_+_30774032 | 0.04 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr21_+_35327589 | 0.03 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr4_+_38937575 | 0.02 |
ENSDART00000150385
|
znf997
|
zinc finger protein 997 |
| chr22_+_1751640 | 0.02 |
ENSDART00000162093
|
znf1169
|
zinc finger protein 1169 |
| chr16_-_37964325 | 0.01 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr20_+_26095530 | 0.01 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr4_+_64921810 | 0.00 |
ENSDART00000157654
|
BX928743.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of gli2a+gli3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 0.9 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 2.2 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 0.8 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 2.2 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 0.8 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 | 1.0 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.6 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.6 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.6 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.3 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 1.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.7 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.2 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.1 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 2.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.7 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 7.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.4 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.5 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 1.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.8 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.4 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.6 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.8 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 2.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 1.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.4 | 1.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 1.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 2.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 2.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.6 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.4 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 1.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.6 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.2 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.1 | 0.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.6 | GO:0030546 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 2.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 2.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |