Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
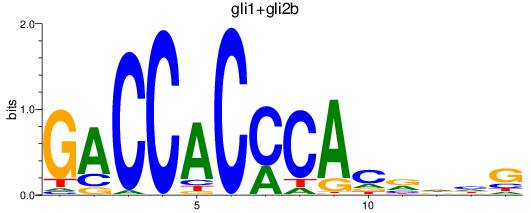
Results for gli1+gli2b
Z-value: 0.95
Transcription factors associated with gli1+gli2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2b
|
ENSDARG00000020884 | GLI family zinc finger 2b |
|
gli1
|
ENSDARG00000101244 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli1 | dr11_v1_chr6_-_59505589_59505589 | -0.61 | 5.4e-03 | Click! |
| gli2b | dr11_v1_chr11_+_43661735_43661735 | -0.61 | 5.8e-03 | Click! |
Activity profile of gli1+gli2b motif
Sorted Z-values of gli1+gli2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_464007 | 3.58 |
ENSDART00000106669
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr20_+_35484070 | 3.21 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr12_-_4756478 | 3.09 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr4_-_8040436 | 2.72 |
ENSDART00000113033
|
si:ch211-240l19.6
|
si:ch211-240l19.6 |
| chr9_-_46399611 | 2.57 |
ENSDART00000164914
ENSDART00000145931 |
lct
si:dkey-79p17.3
|
lactase si:dkey-79p17.3 |
| chr24_-_6158933 | 2.49 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr20_-_25518488 | 2.38 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr14_+_15155684 | 2.37 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr21_-_41873065 | 2.31 |
ENSDART00000014538
|
endou2
|
endonuclease, polyU-specific 2 |
| chr6_+_42918933 | 2.31 |
ENSDART00000064896
|
gnat1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr1_-_21599219 | 2.21 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr17_-_17447899 | 2.21 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr2_+_51783120 | 2.14 |
ENSDART00000177559
|
crygn1
|
crystallin, gamma N1 |
| chr11_+_44300548 | 1.94 |
ENSDART00000191626
|
CABZ01100304.1
|
|
| chr20_+_13969414 | 1.92 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr21_+_10739846 | 1.92 |
ENSDART00000084011
|
cplx4a
|
complexin 4a |
| chr13_+_3954540 | 1.91 |
ENSDART00000092646
|
lrrc73
|
leucine rich repeat containing 73 |
| chr17_+_30587333 | 1.87 |
ENSDART00000156500
|
nhsl1a
|
NHS-like 1a |
| chr19_-_10243148 | 1.82 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr19_-_6988837 | 1.79 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr12_-_31422433 | 1.75 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr23_+_20422661 | 1.74 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr2_+_54482603 | 1.73 |
ENSDART00000130977
ENSDART00000183090 |
mtcl1
|
microtubule crosslinking factor 1 |
| chr2_+_46589798 | 1.73 |
ENSDART00000128457
ENSDART00000145347 |
ephb1
|
EPH receptor B1 |
| chr24_+_3972260 | 1.65 |
ENSDART00000131753
|
pfkpa
|
phosphofructokinase, platelet a |
| chr4_-_17741513 | 1.61 |
ENSDART00000141680
|
mybpc1
|
myosin binding protein C, slow type |
| chr2_-_57707039 | 1.58 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr18_+_25653599 | 1.54 |
ENSDART00000007856
|
fkbp16
|
FK506 binding protein 16 |
| chr5_-_2752573 | 1.46 |
ENSDART00000168121
|
lzts3a
|
leucine zipper, putative tumor suppressor family member 3a |
| chr21_-_41873584 | 1.45 |
ENSDART00000188089
|
endou2
|
endonuclease, polyU-specific 2 |
| chr19_+_49721 | 1.44 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr23_+_28582865 | 1.42 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr14_+_15231097 | 1.41 |
ENSDART00000172430
|
si:dkey-203a12.3
|
si:dkey-203a12.3 |
| chr6_-_11768198 | 1.41 |
ENSDART00000183463
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr20_-_915234 | 1.38 |
ENSDART00000164816
|
cnr1
|
cannabinoid receptor 1 |
| chr2_-_30784502 | 1.36 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr15_+_7992906 | 1.34 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr7_+_29952169 | 1.32 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr12_+_47162456 | 1.32 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr19_+_56351 | 1.29 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr17_-_722218 | 1.28 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr10_-_35542071 | 1.28 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr3_+_19621034 | 1.25 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr6_-_30859656 | 1.23 |
ENSDART00000156235
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr14_-_45702721 | 1.21 |
ENSDART00000165727
|
si:dkey-226n24.1
|
si:dkey-226n24.1 |
| chr1_-_31505144 | 1.17 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr24_-_38574631 | 1.16 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr17_-_50331351 | 1.15 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr2_-_30784198 | 1.13 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr24_-_21587335 | 1.12 |
ENSDART00000091528
|
gpr12
|
G protein-coupled receptor 12 |
| chr17_-_26604946 | 1.12 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr7_+_10701938 | 1.10 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr5_-_34185115 | 1.09 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr1_+_59090743 | 1.09 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr5_+_13472234 | 1.07 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr22_-_38459316 | 1.06 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr20_+_27194833 | 1.06 |
ENSDART00000150072
|
si:dkey-85n7.8
|
si:dkey-85n7.8 |
| chr24_+_39137001 | 1.04 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr19_+_9050852 | 1.04 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr9_+_45227028 | 1.04 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr5_-_17876709 | 1.02 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr7_+_29951997 | 1.01 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr13_+_228045 | 0.99 |
ENSDART00000161091
|
zgc:64201
|
zgc:64201 |
| chr5_-_34185497 | 0.98 |
ENSDART00000146321
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr5_-_25123807 | 0.97 |
ENSDART00000183171
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr8_-_18316482 | 0.96 |
ENSDART00000190156
|
rnf220b
|
ring finger protein 220b |
| chr23_+_26009266 | 0.95 |
ENSDART00000054025
|
si:dkey-78k11.9
|
si:dkey-78k11.9 |
| chr25_-_28384954 | 0.94 |
ENSDART00000073500
|
ptprz1a
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1a |
| chr7_+_4162994 | 0.94 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr7_+_53156810 | 0.90 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr9_-_33749556 | 0.89 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
| chr9_-_46072805 | 0.89 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr13_-_865193 | 0.89 |
ENSDART00000187053
|
AL929536.7
|
|
| chr16_-_31644545 | 0.86 |
ENSDART00000181634
|
CR855311.5
|
|
| chr10_+_5645887 | 0.82 |
ENSDART00000171426
|
pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr14_+_15597049 | 0.80 |
ENSDART00000159732
|
si:dkey-203a12.8
|
si:dkey-203a12.8 |
| chr2_+_30147504 | 0.80 |
ENSDART00000190947
|
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr7_+_29952719 | 0.78 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr15_+_16525126 | 0.77 |
ENSDART00000193455
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr13_-_50002852 | 0.76 |
ENSDART00000099439
|
lyst
|
lysosomal trafficking regulator |
| chr10_-_45379831 | 0.76 |
ENSDART00000186205
|
CABZ01117348.1
|
|
| chr3_-_16068236 | 0.74 |
ENSDART00000157315
|
si:dkey-81l17.6
|
si:dkey-81l17.6 |
| chr9_+_4252839 | 0.74 |
ENSDART00000169740
|
kalrna
|
kalirin RhoGEF kinase a |
| chr14_-_33821515 | 0.73 |
ENSDART00000173218
|
vimr2
|
vimentin-related 2 |
| chr4_+_5132951 | 0.72 |
ENSDART00000103279
|
ccnd2b
|
cyclin D2, b |
| chr17_-_7371564 | 0.71 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr1_+_59090583 | 0.71 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr5_+_69878693 | 0.71 |
ENSDART00000073671
|
ythdc1
|
YTH domain containing 1 |
| chr13_-_9598320 | 0.70 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr4_-_5856200 | 0.69 |
ENSDART00000121936
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr19_-_42651615 | 0.68 |
ENSDART00000123360
|
susd5
|
sushi domain containing 5 |
| chr16_-_12784373 | 0.68 |
ENSDART00000080396
|
foxj2
|
forkhead box J2 |
| chr25_+_26923193 | 0.68 |
ENSDART00000187364
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr14_+_15331486 | 0.68 |
ENSDART00000172077
ENSDART00000183370 ENSDART00000182467 |
SPINK2
si:dkey-203a12.5
|
si:dkey-203a12.4 si:dkey-203a12.5 |
| chr8_-_14121634 | 0.67 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr6_-_31326398 | 0.66 |
ENSDART00000190749
ENSDART00000192789 |
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr8_+_1065458 | 0.64 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr25_+_7461624 | 0.63 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr10_-_15053507 | 0.61 |
ENSDART00000157446
ENSDART00000170441 |
CLDN23
|
si:ch211-95j8.5 |
| chr10_-_39154594 | 0.61 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr25_+_30298377 | 0.60 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr23_+_4226341 | 0.60 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr18_-_1143996 | 0.60 |
ENSDART00000189186
ENSDART00000136140 |
hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr15_+_42431198 | 0.59 |
ENSDART00000189951
ENSDART00000089694 |
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr11_+_2855430 | 0.55 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr4_-_2219705 | 0.54 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr1_+_19434198 | 0.53 |
ENSDART00000012552
|
clockb
|
clock circadian regulator b |
| chr6_+_7123980 | 0.52 |
ENSDART00000179738
ENSDART00000151311 |
si:ch211-237c6.4
|
si:ch211-237c6.4 |
| chr10_+_42691210 | 0.52 |
ENSDART00000193813
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr18_-_16395668 | 0.51 |
ENSDART00000186004
|
mgat4c
|
mgat4 family, member C |
| chr10_-_44411032 | 0.50 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr10_-_15910974 | 0.50 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr6_+_11850359 | 0.47 |
ENSDART00000109552
ENSDART00000188139 ENSDART00000181499 ENSDART00000178269 |
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr21_-_8370500 | 0.47 |
ENSDART00000055328
|
nek6
|
NIMA-related kinase 6 |
| chr4_+_27130412 | 0.44 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr25_-_3087556 | 0.42 |
ENSDART00000193249
|
best1
|
bestrophin 1 |
| chr2_+_54086436 | 0.42 |
ENSDART00000174581
|
CU179656.1
|
|
| chr12_+_30367079 | 0.42 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr1_+_31706668 | 0.42 |
ENSDART00000057879
|
as3mt
|
arsenite methyltransferase |
| chr14_+_25464681 | 0.42 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr23_-_33038423 | 0.41 |
ENSDART00000180539
|
plxna2
|
plexin A2 |
| chr24_-_39518599 | 0.41 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr18_-_17485419 | 0.40 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr19_-_26235219 | 0.39 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr5_+_36439405 | 0.39 |
ENSDART00000102973
|
eda
|
ectodysplasin A |
| chr9_+_56194410 | 0.39 |
ENSDART00000168530
|
LO018176.1
|
|
| chr8_-_7078575 | 0.37 |
ENSDART00000137413
|
si:ch73-233m11.2
|
si:ch73-233m11.2 |
| chr11_+_40831620 | 0.35 |
ENSDART00000160023
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr21_-_36948 | 0.34 |
ENSDART00000181230
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr3_-_25814097 | 0.33 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr14_+_15257658 | 0.33 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr22_+_26665422 | 0.32 |
ENSDART00000164994
|
adcy9
|
adenylate cyclase 9 |
| chr12_-_30338779 | 0.32 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr22_-_3261879 | 0.32 |
ENSDART00000159643
|
gpr35.1
|
G protein-coupled receptor 35, tandem duplicate 1 |
| chr23_+_44049509 | 0.32 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr23_+_18779043 | 0.32 |
ENSDART00000158267
|
emp3b
|
epithelial membrane protein 3b |
| chr15_-_19128705 | 0.31 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr3_+_48561112 | 0.31 |
ENSDART00000159682
|
slc16a5b
|
solute carrier family 16 (monocarboxylate transporter), member 5b |
| chr18_+_5547185 | 0.30 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr1_+_55293424 | 0.30 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr7_+_18075504 | 0.26 |
ENSDART00000173689
|
si:ch73-40a2.1
|
si:ch73-40a2.1 |
| chr20_+_26095530 | 0.25 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr16_-_40373836 | 0.25 |
ENSDART00000134498
|
si:dkey-242e21.3
|
si:dkey-242e21.3 |
| chr16_+_13822137 | 0.24 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr17_-_8562586 | 0.23 |
ENSDART00000154257
ENSDART00000157308 |
fzd3b
|
frizzled class receptor 3b |
| chr12_+_30367371 | 0.23 |
ENSDART00000153364
|
ccdc186
|
si:ch211-225b10.4 |
| chr11_-_21304452 | 0.23 |
ENSDART00000163008
|
RASSF5
|
si:dkey-85p17.3 |
| chr23_+_42304602 | 0.23 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr7_+_31871830 | 0.22 |
ENSDART00000139899
|
mybpc3
|
myosin binding protein C, cardiac |
| chr8_+_28469054 | 0.22 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr20_-_54377933 | 0.19 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr24_+_39518774 | 0.17 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr19_+_13994563 | 0.17 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr11_+_24314148 | 0.16 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr13_-_40754499 | 0.16 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr22_+_38762693 | 0.15 |
ENSDART00000015016
ENSDART00000150187 |
alpi.1
|
alkaline phosphatase, intestinal, tandem duplicate 1 |
| chr12_+_18358082 | 0.14 |
ENSDART00000123315
|
si:dkey-7e14.3
|
si:dkey-7e14.3 |
| chr14_-_15171435 | 0.14 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr16_-_37964325 | 0.14 |
ENSDART00000148801
|
mrap2a
|
melanocortin 2 receptor accessory protein 2a |
| chr25_+_32473433 | 0.14 |
ENSDART00000152326
|
sqor
|
sulfide quinone oxidoreductase |
| chr2_-_12242695 | 0.13 |
ENSDART00000158175
|
gpr158b
|
G protein-coupled receptor 158b |
| chr8_-_43847138 | 0.13 |
ENSDART00000148358
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr21_+_45819662 | 0.11 |
ENSDART00000193362
ENSDART00000184255 |
pitx1
|
paired-like homeodomain 1 |
| chr5_-_18043460 | 0.11 |
ENSDART00000082583
|
rnf215
|
ring finger protein 215 |
| chr5_+_10046643 | 0.10 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr25_-_12982193 | 0.09 |
ENSDART00000159617
|
ccl39.5
|
chemokine (C-C motif) ligand 39, duplicate 5 |
| chr21_+_13389499 | 0.07 |
ENSDART00000151268
|
zgc:113162
|
zgc:113162 |
| chr20_+_2669017 | 0.06 |
ENSDART00000058777
|
slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr5_-_25620594 | 0.04 |
ENSDART00000189346
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr14_+_15430991 | 0.04 |
ENSDART00000158221
|
si:dkey-203a12.5
|
si:dkey-203a12.5 |
| chr16_+_12632428 | 0.03 |
ENSDART00000184600
ENSDART00000180537 |
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr6_-_436658 | 0.02 |
ENSDART00000191515
|
grap2b
|
GRB2-related adaptor protein 2b |
| chr2_-_44199722 | 0.02 |
ENSDART00000140633
ENSDART00000145728 |
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr10_+_40235959 | 0.00 |
ENSDART00000145862
|
gramd1ba
|
GRAM domain containing 1Ba |
Network of associatons between targets according to the STRING database.
First level regulatory network of gli1+gli2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.2 | 0.9 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 2.3 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 1.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 1.4 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.7 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.4 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 1.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.1 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.4 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 1.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 1.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.0 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 3.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.7 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.6 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 2.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.7 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.9 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 2.5 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.6 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.7 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.8 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.7 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 1.6 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.5 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 1.0 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 1.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.5 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.0 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 1.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 1.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.2 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 1.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.3 | 1.4 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.3 | 1.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 2.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.5 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.2 | 1.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.7 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.0 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.3 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.9 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 3.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 1.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 1.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 2.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.0 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 5.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 2.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0048018 | death receptor binding(GO:0005123) receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 2.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.3 | GO:0008013 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) beta-catenin binding(GO:0008013) |
| 0.0 | 1.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.7 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 0.9 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 2.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |