Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
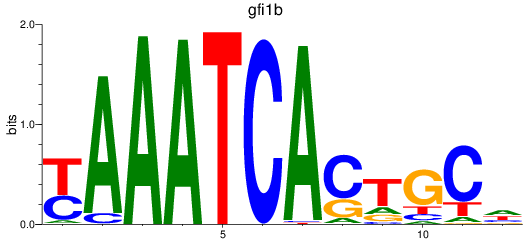
Results for gfi1b
Z-value: 0.47
Transcription factors associated with gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1b
|
ENSDARG00000079947 | growth factor independent 1B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1b | dr11_v1_chr21_-_17296789_17296789 | -0.45 | 5.3e-02 | Click! |
Activity profile of gfi1b motif
Sorted Z-values of gfi1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_32541033 | 0.80 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr23_-_35694461 | 0.78 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr25_+_19999623 | 0.69 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr20_-_26042070 | 0.55 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr17_+_15433671 | 0.55 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr23_-_35694171 | 0.55 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr17_+_15433518 | 0.49 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr19_+_42609132 | 0.47 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr5_-_40734045 | 0.46 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr9_+_30108641 | 0.45 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr10_-_35257458 | 0.43 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr3_-_26109322 | 0.42 |
ENSDART00000113780
|
zgc:162612
|
zgc:162612 |
| chr9_+_19489304 | 0.42 |
ENSDART00000151920
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr19_-_28130658 | 0.40 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr13_-_40398556 | 0.39 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr17_+_24687338 | 0.38 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr7_+_48319916 | 0.36 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr17_-_21066075 | 0.36 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr3_+_23691847 | 0.36 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr25_-_17918536 | 0.36 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr3_-_37148594 | 0.36 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr25_-_15049694 | 0.35 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr5_-_45958838 | 0.34 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr11_-_35171768 | 0.34 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr7_+_22801465 | 0.32 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr11_-_35171162 | 0.32 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr11_+_35050253 | 0.31 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr21_-_33995213 | 0.30 |
ENSDART00000140184
|
EBF1 (1 of many)
|
si:ch211-51e8.2 |
| chr24_-_23716097 | 0.30 |
ENSDART00000084954
ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr25_+_8955530 | 0.29 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr1_+_9860381 | 0.29 |
ENSDART00000054848
|
pmm2
|
phosphomannomutase 2 |
| chr20_-_29864390 | 0.29 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr17_-_33552363 | 0.28 |
ENSDART00000154400
|
tmem121aa
|
transmembrane protein 121Aa |
| chr2_-_15040345 | 0.28 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr7_-_66133786 | 0.28 |
ENSDART00000154961
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr6_-_34220641 | 0.28 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr12_+_8474868 | 0.27 |
ENSDART00000062858
|
adoa
|
2-aminoethanethiol (cysteamine) dioxygenase a |
| chr2_+_9822319 | 0.27 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr8_+_20495889 | 0.27 |
ENSDART00000138794
ENSDART00000021683 |
csnk1g2b
|
casein kinase 1, gamma 2b |
| chr10_-_39153959 | 0.26 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr12_+_27536095 | 0.26 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr5_-_40510397 | 0.26 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr3_-_32818607 | 0.25 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr3_+_51684963 | 0.25 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr18_+_13203831 | 0.24 |
ENSDART00000032151
|
cotl1
|
coactosin-like F-actin binding protein 1 |
| chr3_-_30941362 | 0.24 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr21_+_25068215 | 0.24 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr20_+_50852356 | 0.24 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr13_-_42724645 | 0.24 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr2_+_9821757 | 0.24 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr5_-_24029228 | 0.23 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr6_+_35362225 | 0.23 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr21_+_27370671 | 0.23 |
ENSDART00000009234
ENSDART00000142071 |
rbm14a
|
RNA binding motif protein 14a |
| chr8_-_52715911 | 0.23 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr16_-_45235947 | 0.23 |
ENSDART00000164436
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr19_+_47405867 | 0.23 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr6_+_29791164 | 0.23 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr6_+_27896657 | 0.23 |
ENSDART00000079419
|
cep63
|
centrosomal protein 63 |
| chr24_+_35387517 | 0.22 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr12_+_4220353 | 0.22 |
ENSDART00000133675
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr3_-_36440705 | 0.22 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr25_-_17918810 | 0.22 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr21_+_7605803 | 0.21 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr21_-_17603182 | 0.21 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr23_+_22656477 | 0.21 |
ENSDART00000009337
ENSDART00000133322 |
eno1a
|
enolase 1a, (alpha) |
| chr20_-_28433616 | 0.21 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr9_+_6255682 | 0.21 |
ENSDART00000149827
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr9_+_48007081 | 0.21 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr11_-_15296805 | 0.20 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr18_-_22735002 | 0.20 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr6_+_27897313 | 0.20 |
ENSDART00000128935
|
cep63
|
centrosomal protein 63 |
| chr7_-_35036770 | 0.19 |
ENSDART00000123174
|
galr1b
|
galanin receptor 1b |
| chr25_-_17852944 | 0.19 |
ENSDART00000155169
|
si:ch211-176l24.4
|
si:ch211-176l24.4 |
| chr22_+_10543329 | 0.19 |
ENSDART00000091850
|
atrip
|
ATR interacting protein |
| chr22_+_22437561 | 0.19 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr20_-_28433990 | 0.19 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr3_-_12970418 | 0.18 |
ENSDART00000158747
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr19_+_29798064 | 0.18 |
ENSDART00000167803
ENSDART00000051804 |
marcksl1b
|
MARCKS-like 1b |
| chr11_-_6877973 | 0.18 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr13_-_9119867 | 0.18 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr24_-_4450238 | 0.18 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr8_+_11425048 | 0.18 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr6_-_12912606 | 0.18 |
ENSDART00000164640
|
ical1
|
islet cell autoantigen 1-like |
| chr14_-_16423200 | 0.18 |
ENSDART00000108868
ENSDART00000161793 |
maml1
|
mastermind-like transcriptional coactivator 1 |
| chr4_+_28356606 | 0.18 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr9_-_1965727 | 0.17 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr22_+_16446052 | 0.17 |
ENSDART00000142454
|
si:dkey-121a11.3
|
si:dkey-121a11.3 |
| chr5_+_57773222 | 0.17 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr7_-_28148310 | 0.17 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr9_+_19489514 | 0.17 |
ENSDART00000152032
ENSDART00000114256 ENSDART00000190572 ENSDART00000147571 ENSDART00000151918 ENSDART00000152034 |
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr8_-_1266181 | 0.17 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr9_-_19489264 | 0.17 |
ENSDART00000122894
|
wdr4
|
WD repeat domain 4 |
| chr8_-_51954562 | 0.17 |
ENSDART00000132527
ENSDART00000057315 |
cep78
|
centrosomal protein 78 |
| chr19_+_21919856 | 0.16 |
ENSDART00000187306
ENSDART00000138544 |
galr1a
|
galanin receptor 1a |
| chr7_-_33684632 | 0.16 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr11_+_25044082 | 0.16 |
ENSDART00000123263
|
phf20a
|
PHD finger protein 20, a |
| chr19_+_19600297 | 0.16 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr7_-_25895189 | 0.15 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr14_-_21218891 | 0.15 |
ENSDART00000158294
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr4_-_20235904 | 0.15 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr13_-_43149063 | 0.15 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr13_+_22712406 | 0.15 |
ENSDART00000132847
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr21_-_2935455 | 0.15 |
ENSDART00000163963
|
CABZ01023255.1
|
|
| chr7_-_26408472 | 0.14 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr10_+_9159279 | 0.14 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr5_+_9259971 | 0.14 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr14_-_6943934 | 0.14 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr12_+_21299338 | 0.14 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr24_-_32408404 | 0.14 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr2_+_37836821 | 0.14 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr9_+_6255973 | 0.13 |
ENSDART00000078525
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr13_-_24396199 | 0.13 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr21_-_33995710 | 0.13 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
| chr19_-_12193622 | 0.13 |
ENSDART00000041960
|
ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr5_+_9259574 | 0.13 |
ENSDART00000185560
|
susd1
|
sushi domain containing 1 |
| chr12_-_26022663 | 0.13 |
ENSDART00000166769
|
bmpr1ab
|
bone morphogenetic protein receptor, type IAb |
| chr12_+_27536270 | 0.13 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr7_+_25920792 | 0.13 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr24_-_25096199 | 0.12 |
ENSDART00000185076
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr10_+_35257651 | 0.12 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr20_+_6535176 | 0.12 |
ENSDART00000054652
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr17_+_34805897 | 0.12 |
ENSDART00000137090
ENSDART00000077626 |
id2a
|
inhibitor of DNA binding 2a |
| chr4_-_42242844 | 0.12 |
ENSDART00000163476
|
si:ch211-129p6.2
|
si:ch211-129p6.2 |
| chr7_+_7019911 | 0.12 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr20_+_6535438 | 0.12 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr23_+_23232136 | 0.12 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr7_+_35191220 | 0.12 |
ENSDART00000110552
|
zdhhc1
|
zinc finger, DHHC-type containing 1 |
| chr21_-_131236 | 0.12 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr3_-_39695856 | 0.11 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr3_-_54607166 | 0.11 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr3_-_39696066 | 0.11 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr7_+_69187585 | 0.11 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr11_+_13423776 | 0.10 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr11_+_13424116 | 0.10 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr15_-_1590858 | 0.10 |
ENSDART00000081875
|
nnr
|
nanor |
| chr20_+_28434196 | 0.10 |
ENSDART00000034245
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr25_+_17313568 | 0.10 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr24_+_26402110 | 0.10 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr8_+_23615132 | 0.10 |
ENSDART00000099769
|
ccdc22
|
coiled-coil domain containing 22 |
| chr9_+_44722205 | 0.10 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr17_+_9308425 | 0.10 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr16_-_31824525 | 0.10 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr8_-_15182232 | 0.10 |
ENSDART00000138855
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr12_+_19199735 | 0.09 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr6_+_46431848 | 0.09 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr8_-_49495584 | 0.09 |
ENSDART00000141691
|
opn7d
|
opsin 7, group member d |
| chr12_+_4845448 | 0.09 |
ENSDART00000160666
|
ghrhr2
|
growth hormone releasing hormone receptor 2 |
| chr8_+_37111643 | 0.09 |
ENSDART00000061336
|
ren
|
renin |
| chr4_-_13567387 | 0.09 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr5_-_62306819 | 0.09 |
ENSDART00000168993
|
spata22
|
spermatogenesis associated 22 |
| chr14_-_31151148 | 0.09 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr3_-_41292275 | 0.09 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr3_+_26223376 | 0.09 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr19_-_31584444 | 0.09 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr7_+_39360797 | 0.09 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr20_+_27464721 | 0.09 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr1_+_53221858 | 0.09 |
ENSDART00000191877
|
BX957281.1
|
|
| chr7_+_44445595 | 0.08 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr18_-_18587745 | 0.08 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr17_+_11675362 | 0.08 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr4_-_17642168 | 0.08 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr2_-_30668580 | 0.08 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr24_+_21684189 | 0.08 |
ENSDART00000014696
|
pdx1
|
pancreatic and duodenal homeobox 1 |
| chr13_+_15657911 | 0.08 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr7_+_30201611 | 0.08 |
ENSDART00000075588
|
wdr76
|
WD repeat domain 76 |
| chr21_-_4032650 | 0.07 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr21_+_31253048 | 0.07 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr6_-_3924723 | 0.07 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr17_-_12385308 | 0.07 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr8_-_19395110 | 0.07 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr16_+_23282655 | 0.07 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr17_+_49127003 | 0.07 |
ENSDART00000157360
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr5_-_36924509 | 0.07 |
ENSDART00000025013
|
kptn
|
kaptin (actin binding protein) |
| chr1_+_1805294 | 0.07 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr5_+_43965078 | 0.07 |
ENSDART00000113502
ENSDART00000187143 |
TMEM8B
|
si:dkey-84j12.1 |
| chr16_-_263658 | 0.07 |
ENSDART00000129303
|
slc6a3
|
solute carrier family 6 (neurotransmitter transporter), member 3 |
| chr14_+_3944826 | 0.06 |
ENSDART00000170167
|
lrp2bp
|
LRP2 binding protein |
| chr16_+_10422836 | 0.06 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr5_+_20823409 | 0.06 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr20_-_52642504 | 0.06 |
ENSDART00000138982
ENSDART00000134999 |
si:ch211-221n20.2
|
si:ch211-221n20.2 |
| chr22_-_4407871 | 0.06 |
ENSDART00000162523
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr9_+_13638329 | 0.06 |
ENSDART00000143432
|
als2a
|
amyotrophic lateral sclerosis 2a (juvenile) |
| chr12_-_4220713 | 0.06 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr8_+_21437908 | 0.06 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr11_-_2297832 | 0.06 |
ENSDART00000158266
|
znf740a
|
zinc finger protein 740a |
| chr23_-_9768700 | 0.06 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr4_-_63923523 | 0.06 |
ENSDART00000144330
|
si:dkey-179k24.1
|
si:dkey-179k24.1 |
| chr4_+_13615731 | 0.06 |
ENSDART00000141395
ENSDART00000144310 |
ccdc87
|
coiled-coil domain containing 87 |
| chr20_-_52666150 | 0.06 |
ENSDART00000146716
ENSDART00000134155 |
si:ch211-221n20.4
|
si:ch211-221n20.4 |
| chr8_-_45728620 | 0.06 |
ENSDART00000188795
|
BX088696.1
|
|
| chr16_-_6849754 | 0.05 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr16_-_17560822 | 0.05 |
ENSDART00000089230
|
casp2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr5_-_37871526 | 0.05 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr16_+_26612401 | 0.05 |
ENSDART00000145571
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr13_-_40282770 | 0.05 |
ENSDART00000140875
|
zgc:123010
|
zgc:123010 |
| chr6_+_7466223 | 0.05 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr2_+_34967022 | 0.05 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr6_+_50393047 | 0.05 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr3_-_19367081 | 0.05 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr9_+_37152564 | 0.05 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.4 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.1 | 0.6 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.4 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.2 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.4 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.2 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.2 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.4 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.1 | GO:0048387 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.3 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.2 | GO:0030809 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.4 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0005334 | dopamine transmembrane transporter activity(GO:0005329) dopamine:sodium symporter activity(GO:0005330) norepinephrine transmembrane transporter activity(GO:0005333) norepinephrine:sodium symporter activity(GO:0005334) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |