Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
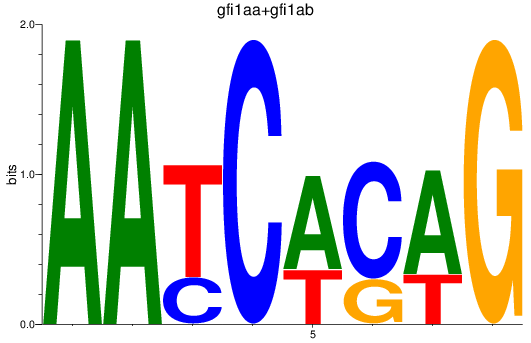
Results for gfi1aa+gfi1ab
Z-value: 0.83
Transcription factors associated with gfi1aa+gfi1ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1aa
|
ENSDARG00000020746 | growth factor independent 1A transcription repressor a |
|
gfi1ab
|
ENSDARG00000044457 | growth factor independent 1A transcription repressor b |
|
gfi1ab
|
ENSDARG00000114140 | growth factor independent 1A transcription repressor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1aa | dr11_v1_chr2_+_10771787_10771787 | -0.33 | 1.7e-01 | Click! |
| gfi1ab | dr11_v1_chr6_-_29007493_29007493 | -0.07 | 7.7e-01 | Click! |
Activity profile of gfi1aa+gfi1ab motif
Sorted Z-values of gfi1aa+gfi1ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_54013457 | 1.09 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr23_-_21471022 | 1.07 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr3_-_1400309 | 1.06 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr16_+_20915319 | 1.05 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr11_-_10456553 | 1.00 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr12_+_27127139 | 1.00 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr23_+_21459263 | 0.93 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr5_-_54714789 | 0.92 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr11_-_10456387 | 0.89 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr9_-_18877597 | 0.89 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr4_+_9508505 | 0.87 |
ENSDART00000080842
|
kitlgb
|
kit ligand b |
| chr24_+_19518570 | 0.87 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr24_+_19518303 | 0.86 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr5_-_19400166 | 0.85 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr6_-_53326421 | 0.84 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr1_+_31573225 | 0.83 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr23_-_21453614 | 0.83 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr14_-_26704829 | 0.82 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr23_+_8797143 | 0.81 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr23_+_21455152 | 0.81 |
ENSDART00000158511
ENSDART00000161321 ENSDART00000160731 ENSDART00000137573 |
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr8_+_15254564 | 0.77 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr25_-_7520937 | 0.77 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr14_-_39074539 | 0.76 |
ENSDART00000030509
|
glra4a
|
glycine receptor, alpha 4a |
| chr15_-_15468326 | 0.76 |
ENSDART00000161192
|
rab34a
|
RAB34, member RAS oncogene family a |
| chr21_+_33187992 | 0.76 |
ENSDART00000162745
ENSDART00000188388 |
BX072577.1
|
|
| chr5_-_54714525 | 0.75 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr19_-_18152942 | 0.75 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr20_-_48485354 | 0.73 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr9_-_3400727 | 0.72 |
ENSDART00000183979
ENSDART00000111386 |
dlx2a
|
distal-less homeobox 2a |
| chr19_-_18152407 | 0.72 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr16_-_20312146 | 0.71 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr3_+_41922114 | 0.70 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_-_16330368 | 0.70 |
ENSDART00000128932
|
epyc
|
epiphycan |
| chr19_-_28130658 | 0.69 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr3_-_32859335 | 0.69 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr4_-_17353100 | 0.68 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr12_-_28848015 | 0.67 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr24_-_8729531 | 0.67 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr1_+_59538755 | 0.66 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr5_-_54712159 | 0.66 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr8_+_28371496 | 0.66 |
ENSDART00000158788
|
klhl12
|
kelch-like family member 12 |
| chr20_-_25645150 | 0.65 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr15_-_4528326 | 0.65 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr23_-_9855627 | 0.65 |
ENSDART00000180159
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr11_-_40519886 | 0.65 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr12_+_27055900 | 0.64 |
ENSDART00000153198
|
fbrs
|
fibrosin |
| chr12_-_8958353 | 0.64 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr21_-_929448 | 0.63 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr19_+_9455218 | 0.63 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr20_+_26940178 | 0.62 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr23_-_1056808 | 0.62 |
ENSDART00000081961
|
zgc:113423
|
zgc:113423 |
| chr19_-_35439237 | 0.62 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr8_+_54055390 | 0.62 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr7_-_34184282 | 0.62 |
ENSDART00000183284
|
smad6a
|
SMAD family member 6a |
| chr16_+_26706519 | 0.61 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr12_+_16087077 | 0.61 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr23_+_36178104 | 0.61 |
ENSDART00000103131
|
hoxc1a
|
homeobox C1a |
| chr12_+_10706772 | 0.61 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr10_-_6427362 | 0.61 |
ENSDART00000166774
|
ca9
|
carbonic anhydrase IX |
| chr9_+_28232522 | 0.60 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr5_-_36948586 | 0.60 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr13_-_16222388 | 0.59 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr20_+_26939742 | 0.59 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr20_-_26491567 | 0.59 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr1_-_45584407 | 0.58 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_+_23563691 | 0.58 |
ENSDART00000142879
|
ncapg
|
non-SMC condensin I complex, subunit G |
| chr22_-_2922053 | 0.58 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr11_+_14104417 | 0.57 |
ENSDART00000059752
ENSDART00000186575 |
med16
|
mediator complex subunit 16 |
| chr20_-_18535502 | 0.57 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr7_-_18598661 | 0.56 |
ENSDART00000182109
|
DTX4
|
si:ch211-119e14.2 |
| chr2_+_49522178 | 0.55 |
ENSDART00000056254
|
stap2a
|
signal transducing adaptor family member 2a |
| chr1_-_38195012 | 0.55 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr13_+_46941930 | 0.55 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr17_+_15433671 | 0.54 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr12_+_25600685 | 0.54 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr22_-_10541372 | 0.53 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr8_+_16738282 | 0.53 |
ENSDART00000134265
ENSDART00000100698 |
ercc8
|
excision repair cross-complementation group 8 |
| chr1_-_50590626 | 0.53 |
ENSDART00000146892
ENSDART00000022733 |
abcg2d
|
ATP-binding cassette, sub-family G (WHITE), member 2d |
| chr2_-_47431205 | 0.53 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr11_+_45343245 | 0.53 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr2_-_29996036 | 0.52 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr7_-_26603743 | 0.52 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr16_-_7443388 | 0.51 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr13_-_45022301 | 0.51 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr8_+_18010978 | 0.51 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr5_-_45958838 | 0.51 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr2_-_48753873 | 0.51 |
ENSDART00000189556
|
CABZ01044731.1
|
|
| chr25_-_15049694 | 0.50 |
ENSDART00000162485
ENSDART00000164384 ENSDART00000165632 ENSDART00000159490 |
pax6a
|
paired box 6a |
| chr3_-_26244256 | 0.50 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr22_+_22437561 | 0.50 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr3_+_23703704 | 0.50 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr13_+_28612313 | 0.50 |
ENSDART00000077383
|
borcs7
|
BLOC-1 related complex subunit 7 |
| chr5_-_23179319 | 0.49 |
ENSDART00000161883
ENSDART00000136260 |
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr22_-_506522 | 0.49 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr9_+_3388099 | 0.49 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr17_-_36936649 | 0.48 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr14_+_30491890 | 0.48 |
ENSDART00000131174
|
fgf20b
|
fibroblast growth factor 20b |
| chr22_+_396840 | 0.48 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr4_-_12477224 | 0.48 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr1_+_34701144 | 0.48 |
ENSDART00000150038
|
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr13_+_51579851 | 0.48 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr18_-_14836600 | 0.48 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr24_+_41690545 | 0.48 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr4_+_4803698 | 0.48 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr3_-_56030117 | 0.48 |
ENSDART00000113030
ENSDART00000157764 |
cep112
|
centrosomal protein 112 |
| chr7_-_16598212 | 0.47 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr13_+_4671698 | 0.47 |
ENSDART00000164617
ENSDART00000128494 ENSDART00000165776 |
pla2g12b
|
phospholipase A2, group XIIB |
| chr2_+_48288461 | 0.47 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr12_+_27536095 | 0.47 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr21_-_26495700 | 0.47 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr3_+_32135037 | 0.47 |
ENSDART00000110490
|
BX511021.1
|
|
| chr13_-_39159810 | 0.47 |
ENSDART00000131508
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr8_-_2591654 | 0.46 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr21_+_30351256 | 0.46 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr2_-_16562505 | 0.46 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr21_+_25068215 | 0.45 |
ENSDART00000167523
ENSDART00000189259 |
dixdc1b
|
DIX domain containing 1b |
| chr6_+_36821621 | 0.45 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr10_-_26274094 | 0.45 |
ENSDART00000108798
|
dchs1b
|
dachsous cadherin-related 1b |
| chr13_+_24263049 | 0.45 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr13_+_30912385 | 0.45 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr17_+_23975762 | 0.44 |
ENSDART00000155941
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr16_+_813780 | 0.44 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr13_+_25433774 | 0.44 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr3_-_27072179 | 0.44 |
ENSDART00000156556
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr8_+_18010568 | 0.44 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr9_-_48214216 | 0.44 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr5_+_17727310 | 0.44 |
ENSDART00000147657
|
fbrsl1
|
fibrosin-like 1 |
| chr22_+_23546926 | 0.44 |
ENSDART00000157940
|
aspm
|
abnormal spindle microtubule assembly |
| chr5_-_35252761 | 0.44 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr24_+_4978055 | 0.43 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr21_+_35215810 | 0.43 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr20_-_20533865 | 0.43 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr16_+_33121260 | 0.43 |
ENSDART00000058472
|
akirin1
|
akirin 1 |
| chr13_-_39160018 | 0.43 |
ENSDART00000168795
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr5_+_9259971 | 0.42 |
ENSDART00000163060
|
susd1
|
sushi domain containing 1 |
| chr2_+_38227264 | 0.42 |
ENSDART00000192707
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr13_-_42536642 | 0.42 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr6_+_40563848 | 0.42 |
ENSDART00000154766
|
si:ch73-15b2.5
|
si:ch73-15b2.5 |
| chr17_-_37795865 | 0.42 |
ENSDART00000025853
|
zfp36l1a
|
zinc finger protein 36, C3H type-like 1a |
| chr15_+_5901970 | 0.42 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr24_+_4977862 | 0.42 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr8_+_37700090 | 0.42 |
ENSDART00000187885
ENSDART00000127633 |
adrb3a
|
adrenoceptor beta 3a |
| chr4_-_16412084 | 0.42 |
ENSDART00000188460
|
dcn
|
decorin |
| chr23_+_2421689 | 0.42 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr23_+_9268083 | 0.41 |
ENSDART00000055054
|
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr12_-_10476448 | 0.41 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr4_+_646154 | 0.41 |
ENSDART00000171138
|
xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr22_-_10541712 | 0.41 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr17_-_43022468 | 0.41 |
ENSDART00000002945
|
NPC2 (1 of many)
|
zgc:193725 |
| chr13_+_43650632 | 0.40 |
ENSDART00000141024
|
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr1_-_20271138 | 0.40 |
ENSDART00000185931
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_+_22258962 | 0.40 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr8_+_25173317 | 0.40 |
ENSDART00000142006
|
gpr61
|
G protein-coupled receptor 61 |
| chr13_+_30912117 | 0.40 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr4_-_9867476 | 0.40 |
ENSDART00000091630
|
fbxo18
|
F-box protein, helicase, 18 |
| chr25_+_27410352 | 0.40 |
ENSDART00000154362
|
pot1
|
protection of telomeres 1 homolog |
| chr2_-_45510699 | 0.40 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr25_-_17918536 | 0.40 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr18_+_16744307 | 0.40 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr18_+_38749547 | 0.40 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr17_-_36936856 | 0.40 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr1_+_14658801 | 0.40 |
ENSDART00000192194
|
CR847936.1
|
|
| chr6_+_48154954 | 0.40 |
ENSDART00000019706
|
phc2b
|
polyhomeotic homolog 2b (Drosophila) |
| chr24_+_21973929 | 0.39 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr8_+_39619087 | 0.39 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr19_+_37120491 | 0.39 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr21_-_10773344 | 0.39 |
ENSDART00000063244
|
grp
|
gastrin-releasing peptide |
| chr19_+_5146460 | 0.39 |
ENSDART00000150740
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr7_-_18470963 | 0.39 |
ENSDART00000173929
ENSDART00000173638 |
znf16l
|
zinc finger protein 16 like |
| chr12_+_27536270 | 0.39 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr8_+_2487250 | 0.38 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr5_+_9259574 | 0.38 |
ENSDART00000185560
|
susd1
|
sushi domain containing 1 |
| chr25_-_23526058 | 0.38 |
ENSDART00000191331
ENSDART00000062930 |
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr6_+_35362225 | 0.38 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr4_-_5652030 | 0.38 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr7_+_40638210 | 0.38 |
ENSDART00000052236
|
mnx1
|
motor neuron and pancreas homeobox 1 |
| chr11_+_14333441 | 0.37 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr10_-_17550239 | 0.37 |
ENSDART00000057513
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr23_+_36095260 | 0.37 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr9_-_43213057 | 0.37 |
ENSDART00000059448
ENSDART00000133589 |
sestd1
|
SEC14 and spectrin domains 1 |
| chr2_+_33926911 | 0.37 |
ENSDART00000109849
ENSDART00000135884 |
kif2c
|
kinesin family member 2C |
| chr19_+_15521997 | 0.37 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr20_-_29864390 | 0.37 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr3_-_32541033 | 0.37 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr1_+_27825980 | 0.37 |
ENSDART00000160524
|
pspc1
|
paraspeckle component 1 |
| chr24_+_40860320 | 0.37 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr6_-_39518489 | 0.36 |
ENSDART00000185446
|
atf1
|
activating transcription factor 1 |
| chr11_-_18017287 | 0.36 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr8_-_4892913 | 0.36 |
ENSDART00000126808
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr6_-_11792152 | 0.36 |
ENSDART00000183403
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr13_-_45022527 | 0.36 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr13_+_31603988 | 0.36 |
ENSDART00000030646
|
six6a
|
SIX homeobox 6a |
| chr3_+_39568290 | 0.35 |
ENSDART00000020741
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr17_-_21066075 | 0.35 |
ENSDART00000078763
ENSDART00000104327 |
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr5_+_43870389 | 0.35 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr9_-_1984604 | 0.35 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr24_+_9881219 | 0.35 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr17_-_11439815 | 0.35 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr10_+_43994471 | 0.34 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr12_+_19191787 | 0.34 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1aa+gfi1ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 1.8 | GO:0055016 | hypochord development(GO:0055016) |
| 0.3 | 2.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 1.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 2.5 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.2 | 0.7 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 0.9 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.2 | 1.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 1.0 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 1.7 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 0.3 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.2 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.4 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 2.8 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.4 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.5 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.6 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.6 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.1 | 0.5 | GO:0048327 | axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 0.1 | 1.0 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 2.0 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 0.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.0 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.3 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.2 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.4 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.8 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.7 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 1.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.4 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.4 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.2 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.1 | GO:2000252 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.6 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.5 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.6 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:2000051 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.0 | 0.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.1 | GO:0033340 | pelvic fin development(GO:0033340) |
| 0.0 | 0.5 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.2 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.0 | 0.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.5 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.6 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 0.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0090505 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.7 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.7 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.7 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0001993 | regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:1900180 | regulation of protein localization to nucleus(GO:1900180) positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.3 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.1 | GO:0001840 | neural plate development(GO:0001840) |
| 0.0 | 0.2 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 1.4 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.3 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.0 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.6 | GO:0072175 | epithelial tube formation(GO:0072175) |
| 0.0 | 0.1 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.4 | GO:0001534 | radial spoke(GO:0001534) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.6 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.2 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.9 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.5 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.6 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 1.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 0.3 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.8 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.4 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 0.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.6 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.5 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.2 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 0.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.1 | 0.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 2.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.2 | GO:1990931 | RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.4 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 19.2 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.1 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.3 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.4 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |