Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for gbx2
Z-value: 0.34
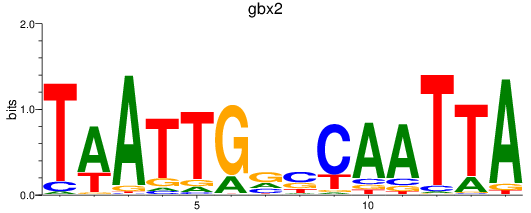
Transcription factors associated with gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gbx2
|
ENSDARG00000002933 | gastrulation brain homeobox 2 |
|
gbx2
|
ENSDARG00000116377 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gbx2 | dr11_v1_chr6_+_16031189_16031189 | 0.16 | 5.2e-01 | Click! |
Activity profile of gbx2 motif
Sorted Z-values of gbx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_14643190 | 0.57 |
ENSDART00000167119
|
znf185
|
zinc finger protein 185 with LIM domain |
| chr5_-_25733745 | 0.45 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr23_+_42254960 | 0.45 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr22_+_9862243 | 0.43 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr6_+_3717613 | 0.42 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr19_+_21820144 | 0.41 |
ENSDART00000181996
|
CU856623.1
|
|
| chr8_+_23826985 | 0.41 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr17_+_1496107 | 0.40 |
ENSDART00000187804
|
LO018430.1
|
|
| chr22_+_9862466 | 0.38 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr10_+_43797130 | 0.38 |
ENSDART00000027242
|
nr2f1b
|
nuclear receptor subfamily 2, group F, member 1b |
| chr15_-_10341048 | 0.35 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr2_-_26720854 | 0.35 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr16_-_7379328 | 0.34 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr22_+_2417105 | 0.32 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr16_+_54875530 | 0.32 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr18_+_17537344 | 0.32 |
ENSDART00000025782
|
nup93
|
nucleoporin 93 |
| chr16_+_5678071 | 0.31 |
ENSDART00000011166
ENSDART00000134198 ENSDART00000131575 |
zgc:158689
|
zgc:158689 |
| chr17_+_10593398 | 0.30 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr11_+_31323746 | 0.30 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr15_-_16177603 | 0.30 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr9_-_30555725 | 0.29 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr3_+_28581397 | 0.29 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr7_-_33684632 | 0.28 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr24_-_26995164 | 0.27 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr24_-_20359350 | 0.27 |
ENSDART00000109223
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr16_+_27593698 | 0.26 |
ENSDART00000145203
|
si:ch211-197h24.8
|
si:ch211-197h24.8 |
| chr20_+_18741089 | 0.26 |
ENSDART00000183218
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr7_+_20260172 | 0.25 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr16_+_16968682 | 0.25 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr7_+_46019780 | 0.25 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr13_-_330004 | 0.24 |
ENSDART00000093149
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr17_+_24318753 | 0.24 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr22_-_9861531 | 0.24 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr18_+_7639401 | 0.23 |
ENSDART00000092416
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr15_-_45110011 | 0.23 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr21_-_40348790 | 0.22 |
ENSDART00000178123
|
CR847523.1
|
|
| chr24_+_38301080 | 0.22 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr22_+_2512154 | 0.21 |
ENSDART00000097363
|
zgc:173726
|
zgc:173726 |
| chr12_-_34713690 | 0.20 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr16_+_16969060 | 0.20 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr22_-_14272699 | 0.19 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr12_-_35386910 | 0.18 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr6_-_42369186 | 0.18 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr12_+_5977777 | 0.18 |
ENSDART00000152302
|
si:ch211-131k2.2
|
si:ch211-131k2.2 |
| chr10_-_1788376 | 0.18 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr23_-_30727596 | 0.17 |
ENSDART00000060193
|
thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr5_+_27897504 | 0.17 |
ENSDART00000130936
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr18_-_15932704 | 0.16 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr7_-_46019756 | 0.16 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr12_+_22560067 | 0.16 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr3_-_26921475 | 0.16 |
ENSDART00000130281
|
ciita
|
class II, major histocompatibility complex, transactivator |
| chr9_-_3934963 | 0.15 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr11_+_24340166 | 0.15 |
ENSDART00000188719
|
rbm39a
|
RNA binding motif protein 39a |
| chr8_+_21229718 | 0.15 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr7_+_53755054 | 0.15 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr5_-_7834582 | 0.15 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr18_+_26895994 | 0.15 |
ENSDART00000098347
|
ch25hl1.2
|
cholesterol 25-hydroxylase like 1, tandem duplicate 2 |
| chr10_-_43113731 | 0.14 |
ENSDART00000138099
|
tmem167a
|
transmembrane protein 167A |
| chr9_+_23003208 | 0.14 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr7_+_59677273 | 0.14 |
ENSDART00000039535
ENSDART00000132044 |
trmt44
|
tRNA methyltransferase 44 homolog |
| chr23_-_333457 | 0.13 |
ENSDART00000114486
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr16_-_26855936 | 0.13 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr22_-_36530902 | 0.13 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr24_-_16904234 | 0.12 |
ENSDART00000192879
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr24_-_33801668 | 0.12 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr11_+_6819050 | 0.12 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr23_+_44349252 | 0.12 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr19_-_27336092 | 0.12 |
ENSDART00000176885
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr10_-_14929630 | 0.12 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr8_+_23105117 | 0.11 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr8_+_20140321 | 0.11 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr3_-_21037840 | 0.11 |
ENSDART00000002393
|
rundc3aa
|
RUN domain containing 3Aa |
| chr10_-_11761927 | 0.10 |
ENSDART00000138748
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr14_-_33351103 | 0.10 |
ENSDART00000010019
|
upf3b
|
UPF3B, regulator of nonsense mediated mRNA decay |
| chr11_-_12158412 | 0.10 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr19_-_10330778 | 0.09 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr16_-_35579200 | 0.09 |
ENSDART00000162172
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr8_+_18010568 | 0.08 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr18_+_16750080 | 0.08 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr22_-_619711 | 0.08 |
ENSDART00000192778
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr5_-_69923241 | 0.08 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr20_-_49889111 | 0.08 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr19_-_15855427 | 0.07 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr19_-_20430892 | 0.07 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr11_-_2632747 | 0.07 |
ENSDART00000008133
|
si:ch211-160o17.2
|
si:ch211-160o17.2 |
| chr13_-_33654931 | 0.07 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr18_-_16937008 | 0.06 |
ENSDART00000100117
ENSDART00000022640 ENSDART00000136541 |
znf143b
|
zinc finger protein 143b |
| chr10_-_39052264 | 0.06 |
ENSDART00000144036
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr5_+_57773222 | 0.06 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr4_-_5018705 | 0.05 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr9_-_9671244 | 0.05 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr2_+_41926707 | 0.04 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr9_+_55154414 | 0.04 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr6_+_9191981 | 0.03 |
ENSDART00000150916
|
zgc:112392
|
zgc:112392 |
| chr14_+_35464994 | 0.03 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr13_+_733027 | 0.03 |
ENSDART00000149547
|
nrxn1b
|
neurexin 1b |
| chr3_-_10739625 | 0.03 |
ENSDART00000156144
|
zgc:112965
|
zgc:112965 |
| chr25_-_31702970 | 0.02 |
ENSDART00000153586
|
lamb4
|
laminin, beta 4 |
| chr11_+_16216909 | 0.02 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr12_-_20665164 | 0.02 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr17_+_49281597 | 0.01 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr8_-_25729542 | 0.01 |
ENSDART00000078149
ENSDART00000085779 |
foxp3a
|
forkhead box P3a |
| chr8_+_44478294 | 0.00 |
ENSDART00000006898
|
si:ch73-211l2.3
|
si:ch73-211l2.3 |
| chr15_+_34934568 | 0.00 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gbx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.3 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.4 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.2 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |