Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
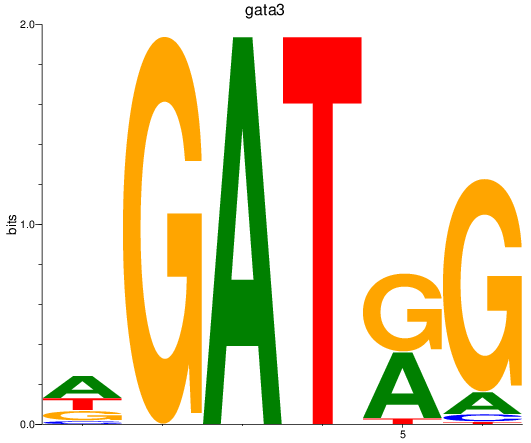
Results for gata3
Z-value: 2.15
Transcription factors associated with gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata3
|
ENSDARG00000016526 | GATA binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata3 | dr11_v1_chr4_-_25064510_25064510 | 0.24 | 3.2e-01 | Click! |
Activity profile of gata3 motif
Sorted Z-values of gata3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_43079161 | 11.01 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr20_-_47550577 | 10.94 |
ENSDART00000187361
|
CABZ01053323.1
|
|
| chr7_+_56615554 | 10.90 |
ENSDART00000098430
|
dpep1
|
dipeptidase 1 |
| chr5_-_7199998 | 10.26 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr11_+_36989696 | 9.76 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr19_+_42219165 | 8.93 |
ENSDART00000163192
|
CU896644.1
|
|
| chr7_+_13382852 | 8.27 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr18_-_22753637 | 7.97 |
ENSDART00000181589
ENSDART00000009912 |
hsf4
|
heat shock transcription factor 4 |
| chr9_-_35557397 | 7.50 |
ENSDART00000100681
|
ncam2
|
neural cell adhesion molecule 2 |
| chr6_+_2174082 | 7.40 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr22_+_15507218 | 7.12 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr17_+_40684 | 7.08 |
ENSDART00000164231
|
CABZ01111555.1
|
|
| chr17_+_50524573 | 6.28 |
ENSDART00000187974
|
CR382385.1
|
|
| chr22_+_1137997 | 6.27 |
ENSDART00000158017
ENSDART00000183226 |
si:ch211-103a14.5
|
si:ch211-103a14.5 |
| chr9_+_45605410 | 5.91 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr1_-_14258409 | 5.85 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr2_+_51796441 | 5.80 |
ENSDART00000165151
|
crygn1
|
crystallin, gamma N1 |
| chr14_-_1565317 | 5.75 |
ENSDART00000169496
|
CABZ01064248.1
|
|
| chr23_-_41762956 | 5.43 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr8_-_3716734 | 4.98 |
ENSDART00000172966
|
bicdl1
|
BICD family like cargo adaptor 1 |
| chr8_-_6825588 | 4.78 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr24_-_29822913 | 4.77 |
ENSDART00000160929
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr4_-_4932619 | 4.70 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr7_-_72067475 | 4.65 |
ENSDART00000017763
|
LO018380.1
|
|
| chr19_-_36675023 | 4.62 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr16_-_29437373 | 4.44 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr25_-_17403598 | 4.29 |
ENSDART00000104216
|
cyp2x9
|
cytochrome P450, family 2, subfamily X, polypeptide 9 |
| chr20_+_9828769 | 4.20 |
ENSDART00000160480
ENSDART00000053844 ENSDART00000080691 |
stxbp6l
|
syntaxin binding protein 6 (amisyn), like |
| chr2_+_33541928 | 4.19 |
ENSDART00000162852
|
BX548164.1
|
|
| chr7_+_15659280 | 4.10 |
ENSDART00000173414
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr9_-_51370293 | 4.07 |
ENSDART00000084806
|
slc4a10b
|
solute carrier family 4, sodium bicarbonate transporter, member 10b |
| chr21_-_43022048 | 4.06 |
ENSDART00000138329
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr21_+_24287403 | 4.01 |
ENSDART00000111169
|
cadm1a
|
cell adhesion molecule 1a |
| chr19_-_12322356 | 3.99 |
ENSDART00000016128
|
ncaldb
|
neurocalcin delta b |
| chr5_+_44654535 | 3.95 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr11_+_34824099 | 3.87 |
ENSDART00000037017
ENSDART00000146944 |
slc38a3a
|
solute carrier family 38, member 3a |
| chr10_-_10018120 | 3.84 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr23_+_2542158 | 3.79 |
ENSDART00000182197
|
LO017835.1
|
|
| chr25_-_36827491 | 3.70 |
ENSDART00000170941
|
CU929365.1
|
|
| chr17_+_20607487 | 3.69 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr21_-_3844322 | 3.59 |
ENSDART00000166652
|
st6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr2_+_53204750 | 3.56 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr11_-_2700397 | 3.53 |
ENSDART00000082511
|
si:ch211-160o17.6
|
si:ch211-160o17.6 |
| chr23_-_41762797 | 3.53 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr21_+_30043054 | 3.50 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr10_-_9192450 | 3.48 |
ENSDART00000139783
|
si:dkeyp-41f9.4
|
si:dkeyp-41f9.4 |
| chr19_-_43819582 | 3.44 |
ENSDART00000160879
ENSDART00000075902 |
klhl43
|
kelch-like family member 43 |
| chr7_-_24699985 | 3.43 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr17_-_8173380 | 3.40 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr12_+_13256415 | 3.39 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr24_-_24281792 | 3.33 |
ENSDART00000146482
ENSDART00000018420 |
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr20_+_18163355 | 3.29 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr22_+_35068046 | 3.27 |
ENSDART00000161660
ENSDART00000169573 |
si:ch73-173h19.3
|
si:ch73-173h19.3 |
| chr18_+_8805575 | 3.24 |
ENSDART00000137098
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr16_-_16649764 | 3.23 |
ENSDART00000014439
|
tnxba
|
tenascin XBa |
| chr15_+_40008370 | 3.12 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr15_-_29598679 | 3.12 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr15_-_29598444 | 3.11 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr24_-_24282427 | 3.09 |
ENSDART00000123299
|
pdha1b
|
pyruvate dehydrogenase E1 alpha 1 subunit b |
| chr13_-_46991577 | 3.08 |
ENSDART00000114748
|
vip
|
vasoactive intestinal peptide |
| chr11_+_34824262 | 3.07 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
| chr25_+_336503 | 3.05 |
ENSDART00000160395
|
CU929262.1
|
|
| chr3_+_23247325 | 3.05 |
ENSDART00000114190
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr11_+_1628538 | 3.04 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr13_-_31544365 | 3.04 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr20_-_44557037 | 2.98 |
ENSDART00000140995
|
mfsd2b
|
major facilitator superfamily domain containing 2B |
| chr4_+_6643421 | 2.97 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr6_-_15604157 | 2.96 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr11_-_1131569 | 2.95 |
ENSDART00000173228
|
slc6a11b
|
solute carrier family 6 (neurotransmitter transporter), member 11b |
| chr20_+_22130284 | 2.82 |
ENSDART00000025575
|
clocka
|
clock circadian regulator a |
| chr22_+_1092479 | 2.81 |
ENSDART00000170119
|
guca1e
|
guanylate cyclase activator 1e |
| chr23_-_16692312 | 2.77 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr15_+_31735931 | 2.76 |
ENSDART00000185681
ENSDART00000149137 |
rxfp2b
|
relaxin/insulin-like family peptide receptor 2b |
| chr7_-_13381129 | 2.71 |
ENSDART00000164326
|
si:ch73-119p20.1
|
si:ch73-119p20.1 |
| chr4_+_4267451 | 2.69 |
ENSDART00000192069
|
ano2
|
anoctamin 2 |
| chr14_+_50770537 | 2.67 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr17_+_45305645 | 2.64 |
ENSDART00000172488
|
capn3a
|
calpain 3a, (p94) |
| chr10_-_2527342 | 2.64 |
ENSDART00000184168
|
CU856539.1
|
|
| chr25_-_19433244 | 2.59 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr5_-_2282256 | 2.58 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr22_+_997838 | 2.57 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr2_-_42705284 | 2.52 |
ENSDART00000187160
|
myo10
|
myosin X |
| chr1_-_54765262 | 2.52 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr15_-_33896159 | 2.44 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr18_-_6460102 | 2.42 |
ENSDART00000137037
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr7_+_71955486 | 2.37 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr16_-_52736549 | 2.34 |
ENSDART00000146607
|
azin1a
|
antizyme inhibitor 1a |
| chr6_-_15604417 | 2.31 |
ENSDART00000157817
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr14_+_36497250 | 2.27 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr4_-_19884440 | 2.24 |
ENSDART00000182979
ENSDART00000105967 |
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr8_+_19621511 | 2.21 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr16_-_13730152 | 2.18 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr1_-_21909608 | 2.16 |
ENSDART00000139937
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr18_-_42785469 | 2.13 |
ENSDART00000024768
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr20_-_7648325 | 2.13 |
ENSDART00000186541
|
CR848049.1
|
|
| chr12_-_32421046 | 2.11 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr15_+_9294620 | 2.11 |
ENSDART00000133588
ENSDART00000140009 |
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr4_-_19883985 | 2.11 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr16_+_52512025 | 2.09 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr8_-_12403077 | 2.09 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr15_+_6109861 | 2.03 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr6_+_21740672 | 2.02 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr8_-_22698651 | 2.02 |
ENSDART00000181411
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr3_-_13146631 | 2.01 |
ENSDART00000172460
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr23_+_42434348 | 2.00 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr20_-_43775495 | 2.00 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr9_-_44953349 | 1.99 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr17_+_5768608 | 1.98 |
ENSDART00000157039
|
rp1l1a
|
retinitis pigmentosa 1-like 1a |
| chr19_+_43314700 | 1.97 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr11_-_36020005 | 1.97 |
ENSDART00000031993
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr7_+_49865049 | 1.97 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr3_-_21242460 | 1.96 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr6_+_51773873 | 1.96 |
ENSDART00000156516
|
tmem74b
|
transmembrane protein 74B |
| chr17_-_37214196 | 1.95 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr18_+_16963881 | 1.93 |
ENSDART00000147583
|
si:ch211-242e8.1
|
si:ch211-242e8.1 |
| chr22_+_635813 | 1.92 |
ENSDART00000179067
|
CU856139.1
|
|
| chr15_-_27972474 | 1.89 |
ENSDART00000162753
|
CR391930.1
|
|
| chr22_+_15438872 | 1.89 |
ENSDART00000139800
|
gpc5b
|
glypican 5b |
| chr22_-_2959005 | 1.89 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr20_-_34754617 | 1.88 |
ENSDART00000148066
|
znf395b
|
zinc finger protein 395b |
| chr20_-_52902693 | 1.88 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr17_-_5732299 | 1.88 |
ENSDART00000017244
|
xkr6a
|
XK, Kell blood group complex subunit-related family, member 6a |
| chr21_-_41588129 | 1.86 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr2_-_3158919 | 1.80 |
ENSDART00000098394
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr12_+_5189776 | 1.79 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr2_+_35854242 | 1.79 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr17_-_26604549 | 1.79 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr10_+_3127809 | 1.79 |
ENSDART00000185500
|
ckmt2a
|
creatine kinase, mitochondrial 2a (sarcomeric) |
| chr7_-_36096582 | 1.79 |
ENSDART00000188507
|
CR848715.1
|
|
| chr7_+_26326462 | 1.77 |
ENSDART00000173515
|
zanl
|
zonadhesin, like |
| chr21_+_43561650 | 1.75 |
ENSDART00000085071
|
gpr185a
|
G protein-coupled receptor 185 a |
| chr8_-_8446668 | 1.75 |
ENSDART00000132700
|
cdk16
|
cyclin-dependent kinase 16 |
| chr22_+_15438513 | 1.73 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr1_-_23557877 | 1.70 |
ENSDART00000145942
|
fam184b
|
family with sequence similarity 184, member B |
| chr15_-_5016039 | 1.70 |
ENSDART00000156458
|
defbl3
|
defensin, beta-like 3 |
| chr3_-_57779801 | 1.67 |
ENSDART00000074285
|
syngr2a
|
synaptogyrin 2a |
| chr12_+_13244149 | 1.66 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr2_+_37207461 | 1.65 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr7_-_48173440 | 1.63 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr1_-_55238610 | 1.60 |
ENSDART00000110818
|
FO704915.1
|
|
| chr12_+_16106459 | 1.60 |
ENSDART00000114174
|
muc13b
|
mucin 13b, cell surface associated |
| chr4_+_61995745 | 1.58 |
ENSDART00000171539
|
CT990567.1
|
|
| chr16_-_36834505 | 1.54 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr18_+_44769211 | 1.53 |
ENSDART00000177181
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr6_-_55585423 | 1.53 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr7_-_13884610 | 1.52 |
ENSDART00000006897
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr1_-_52128425 | 1.50 |
ENSDART00000149939
|
rad23aa
|
RAD23 homolog A, nucleotide excision repair protein a |
| chr16_-_35532937 | 1.50 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr8_-_33154677 | 1.50 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr7_+_10610791 | 1.49 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr20_-_47422469 | 1.49 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr6_-_37403444 | 1.48 |
ENSDART00000136314
|
cthl
|
cystathionase (cystathionine gamma-lyase), like |
| chr25_+_4760489 | 1.47 |
ENSDART00000167399
|
FP245455.1
|
|
| chr17_-_37395460 | 1.46 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr17_+_15033822 | 1.46 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr3_-_33901483 | 1.46 |
ENSDART00000144774
ENSDART00000138765 |
cacna1aa
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a |
| chr8_+_26859639 | 1.46 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr25_+_33063762 | 1.44 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr20_-_52939501 | 1.44 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr6_-_609880 | 1.43 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr2_+_30144979 | 1.43 |
ENSDART00000129365
ENSDART00000130344 |
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr8_-_44835944 | 1.43 |
ENSDART00000185033
|
FO082780.1
|
|
| chr25_-_11088839 | 1.43 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr11_+_40032790 | 1.42 |
ENSDART00000158809
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr17_+_28882977 | 1.40 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr5_+_9360394 | 1.38 |
ENSDART00000124642
|
FP236810.2
|
|
| chr10_+_37182626 | 1.38 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr16_-_22683386 | 1.37 |
ENSDART00000077978
|
s100t
|
S100 calcium binding protein T |
| chr5_-_54197084 | 1.36 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr16_-_45178430 | 1.35 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr2_+_9552456 | 1.35 |
ENSDART00000056896
|
dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr1_-_55044256 | 1.34 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr6_+_45346808 | 1.34 |
ENSDART00000186327
|
LO017951.1
|
|
| chr12_-_20362041 | 1.33 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr17_-_20897250 | 1.33 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr1_+_1941031 | 1.33 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr8_-_21268303 | 1.31 |
ENSDART00000067211
|
gpr37l1b
|
G protein-coupled receptor 37 like 1b |
| chr10_-_19497914 | 1.30 |
ENSDART00000132084
|
si:ch211-127i16.2
|
si:ch211-127i16.2 |
| chr23_+_41831224 | 1.30 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr24_-_29997145 | 1.28 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr9_-_8670158 | 1.28 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr10_-_29809105 | 1.27 |
ENSDART00000162231
|
si:ch73-261i21.5
|
si:ch73-261i21.5 |
| chr17_-_17447899 | 1.27 |
ENSDART00000156928
ENSDART00000109034 |
nrxn3a
|
neurexin 3a |
| chr15_-_30984557 | 1.27 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr5_-_32308526 | 1.26 |
ENSDART00000193758
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr16_+_14029283 | 1.25 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr9_+_30274637 | 1.25 |
ENSDART00000079095
|
rpgra
|
retinitis pigmentosa GTPase regulator a |
| chr22_-_263117 | 1.24 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr10_-_44241119 | 1.24 |
ENSDART00000164480
|
CABZ01087091.1
|
|
| chr22_-_237651 | 1.23 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr11_+_29975830 | 1.23 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr23_+_2778813 | 1.22 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr17_+_23554932 | 1.22 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr6_-_46875310 | 1.22 |
ENSDART00000154442
|
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr15_+_42933236 | 1.21 |
ENSDART00000167763
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr10_+_44903676 | 1.21 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr7_+_50053339 | 1.20 |
ENSDART00000174308
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr20_+_48413712 | 1.20 |
ENSDART00000159983
|
CABZ01059119.1
|
|
| chr5_-_68641353 | 1.20 |
ENSDART00000048432
|
dlg4a
|
discs, large homolog 4a (Drosophila) |
| chr15_+_32710300 | 1.20 |
ENSDART00000167857
|
postnb
|
periostin, osteoblast specific factor b |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 2.1 | 8.3 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 2.0 | 9.8 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 1.7 | 7.0 | GO:0006867 | asparagine transport(GO:0006867) |
| 1.2 | 5.0 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 1.1 | 6.4 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.9 | 3.4 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.8 | 3.1 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.8 | 3.1 | GO:0014060 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.7 | 5.9 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.5 | 2.0 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.5 | 2.0 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.5 | 2.0 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.5 | 1.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.5 | 2.3 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.5 | 1.8 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.4 | 2.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 1.2 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 5.5 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.4 | 2.3 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.4 | 1.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.4 | 3.3 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 4.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 1.0 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 0.9 | GO:0009750 | response to fructose(GO:0009750) |
| 0.3 | 0.9 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.3 | 2.9 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.3 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 0.6 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.3 | 1.7 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.3 | 3.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 1.0 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 5.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 0.5 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.2 | 2.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.2 | 0.7 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.2 | 1.4 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 6.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 0.9 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 2.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 1.4 | GO:0090134 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 1.1 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.2 | 1.5 | GO:0019346 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.2 | 2.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.9 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.2 | 0.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.9 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.2 | 0.4 | GO:0032616 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.2 | 0.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 0.7 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 1.1 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 1.4 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.2 | 2.8 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.2 | 4.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.2 | 0.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.2 | 3.7 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 0.8 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.2 | 0.5 | GO:0032847 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.2 | 1.0 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.2 | 7.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.2 | 1.8 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 0.5 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.2 | 0.8 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 1.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 3.7 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 3.0 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.2 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.5 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 1.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 0.1 | GO:0032768 | regulation of monooxygenase activity(GO:0032768) positive regulation of monooxygenase activity(GO:0032770) |
| 0.1 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.1 | 0.6 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 10.1 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.1 | 1.8 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.5 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.7 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.4 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.1 | 0.7 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 3.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.9 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 1.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 1.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 1.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 5.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.5 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.5 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 2.7 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.9 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.5 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.3 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.8 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.9 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 3.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.3 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.6 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 1.7 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.1 | 0.4 | GO:0045988 | regulation of twitch skeletal muscle contraction(GO:0014724) fast-twitch skeletal muscle fiber contraction(GO:0031443) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.9 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 4.9 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 0.7 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 2.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.0 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 0.6 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.5 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.6 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.7 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 2.7 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 1.6 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.4 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.5 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 15.0 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 2.7 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 2.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.5 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.1 | 0.3 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 1.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 2.3 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 0.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 1.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 5.0 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.8 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 0.6 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 3.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 4.7 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 3.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.7 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.9 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 0.6 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.2 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.1 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 11.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 5.4 | GO:0001666 | response to hypoxia(GO:0001666) |
| 0.1 | 0.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.2 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.6 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.1 | GO:0010543 | regulation of platelet activation(GO:0010543) regulation of platelet aggregation(GO:0090330) |
| 0.1 | 0.4 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.3 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.6 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.1 | 2.5 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.8 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.1 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.2 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.7 | GO:0030816 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 2.1 | GO:1903038 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 5.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.5 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.0 | 0.2 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.4 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.8 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.3 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.8 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.5 | GO:1990573 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 4.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 2.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.2 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.5 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 1.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.8 | GO:0031529 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.8 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.3 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 1.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.5 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 1.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.5 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.9 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.3 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 3.3 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 1.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 1.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.0 | 3.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.3 | GO:0051384 | response to corticosteroid(GO:0031960) response to glucocorticoid(GO:0051384) cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 2.9 | GO:0034765 | regulation of transmembrane transport(GO:0034762) regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 1.4 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.5 | GO:0010469 | regulation of receptor activity(GO:0010469) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 1.5 | GO:0007270 | neuron-neuron synaptic transmission(GO:0007270) synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.9 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 4.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.0 | 0.3 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.3 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.0 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 1.1 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.6 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.0 | GO:0060043 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 4.0 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) |
| 0.0 | 0.4 | GO:0071222 | cellular response to biotic stimulus(GO:0071216) cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 1.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) tendon formation(GO:0035992) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.3 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.3 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.8 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.6 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.1 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.0 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.3 | GO:0006885 | regulation of pH(GO:0006885) regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.0 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.4 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.6 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.4 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.7 | 2.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.7 | 2.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.6 | 4.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.6 | 1.8 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.6 | 7.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 2.8 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.5 | 2.5 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 10.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 1.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.3 | 13.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 2.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 2.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 5.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 1.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 3.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.2 | 1.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 3.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 6.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 8.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 4.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 2.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.6 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.1 | 0.9 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 2.0 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 4.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 4.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 4.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.5 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 8.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.5 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 4.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.3 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) AMPA glutamate receptor complex(GO:0032281) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 2.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 8.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.1 | 4.9 | GO:0044297 | cell body(GO:0044297) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.1 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.7 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.2 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.0 | 0.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 2.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 6.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.7 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 2.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 14.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.0 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 17.9 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.9 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 12.5 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 1.9 | 11.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 1.3 | 6.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 1.3 | 7.6 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 1.2 | 4.8 | GO:0004135 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 1.2 | 9.4 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 1.2 | 7.0 | GO:0015182 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 1.1 | 3.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 1.0 | 6.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.9 | 2.7 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.8 | 8.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.7 | 5.6 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 4.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 3.0 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.5 | 2.9 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 5.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 3.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 4.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 5.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.4 | 2.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.4 | 2.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.4 | 2.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.4 | 1.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.4 | 1.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.3 | 5.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 1.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 1.4 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.3 | 1.0 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 2.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.9 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.2 | 0.9 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.2 | 1.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 3.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 1.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.3 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.2 | 4.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.8 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 1.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.8 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 0.6 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.2 | 1.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.9 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 0.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 1.8 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 1.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 2.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 2.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 0.5 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 8.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 1.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.2 | 0.8 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.1 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.2 | 0.9 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.2 | 0.5 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.2 | 2.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 3.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.7 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.4 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 3.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.4 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.1 | 0.5 | GO:0061133 | peptidase activator activity(GO:0016504) endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.6 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 1.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 4.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.7 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 3.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 9.1 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.4 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 2.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 3.1 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 6.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.5 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 5.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.7 | GO:0051393 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.4 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.4 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 2.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.8 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 2.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.8 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.5 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 1.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.7 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.1 | 0.7 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.5 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.6 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 5.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 1.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 2.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.7 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 3.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.8 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.2 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 2.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.8 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.9 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 13.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 12.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 3.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 3.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.6 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 1.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.6 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 6.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 4.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.0 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 10.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 4.5 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.2 | 2.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 0.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 2.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.6 | 3.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.4 | 4.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.4 | 3.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 3.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 8.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.3 | 4.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 1.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 1.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.4 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 2.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.8 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 4.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.9 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.3 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.5 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 4.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 1.7 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |