Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
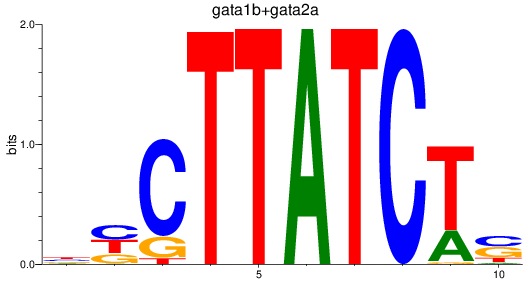
Results for gata1b+gata2a
Z-value: 2.89
Transcription factors associated with gata1b+gata2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata1b
|
ENSDARG00000059130 | GATA binding protein 1b |
|
gata2a
|
ENSDARG00000059327 | GATA binding protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata2a | dr11_v1_chr11_-_3860534_3860534 | -0.77 | 1.1e-04 | Click! |
| gata1b | dr11_v1_chr8_-_7475917_7475917 | 0.51 | 2.5e-02 | Click! |
Activity profile of gata1b+gata2a motif
Sorted Z-values of gata1b+gata2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_32421046 | 15.44 |
ENSDART00000075567
|
enpp7.1
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 1 |
| chr5_-_30620625 | 14.77 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr7_+_35075847 | 14.21 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr2_-_51794472 | 14.12 |
ENSDART00000186652
|
BX908782.3
|
|
| chr16_-_36834505 | 13.21 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr19_+_8606883 | 12.60 |
ENSDART00000054469
ENSDART00000185264 |
s100a10a
|
S100 calcium binding protein A10a |
| chr14_+_36738069 | 11.12 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr18_-_42830563 | 9.46 |
ENSDART00000191488
|
ttc36
|
tetratricopeptide repeat domain 36 |
| chr12_-_3903886 | 9.08 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr21_-_40880317 | 9.02 |
ENSDART00000100054
ENSDART00000137696 |
elnb
|
elastin b |
| chr1_+_9954489 | 9.01 |
ENSDART00000005895
ENSDART00000183003 |
pdia2
|
protein disulfide isomerase family A, member 2 |
| chr20_+_33981946 | 8.78 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr14_+_32839535 | 7.94 |
ENSDART00000168975
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr8_-_40555340 | 7.75 |
ENSDART00000163348
|
NPC1L1
|
NPC1 like intracellular cholesterol transporter 1 |
| chr6_-_609880 | 7.29 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr5_+_26248380 | 6.51 |
ENSDART00000079049
|
si:ch211-214j8.1
|
si:ch211-214j8.1 |
| chr14_+_17382803 | 6.39 |
ENSDART00000040383
|
si:ch211-255i20.3
|
si:ch211-255i20.3 |
| chr25_-_7974494 | 6.21 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr19_+_37509638 | 6.16 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr13_+_10945337 | 6.16 |
ENSDART00000091845
|
abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr7_-_73752955 | 6.13 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr16_+_5612547 | 6.12 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr8_+_26268100 | 6.03 |
ENSDART00000138882
|
slc26a6
|
solute carrier family 26, member 6 |
| chr2_+_39021282 | 6.01 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr4_+_7508316 | 5.91 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr15_-_43164591 | 5.88 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr3_+_16449490 | 5.61 |
ENSDART00000141467
|
kcnj12b
|
potassium inwardly-rectifying channel, subfamily J, member 12b |
| chr3_-_33395233 | 5.36 |
ENSDART00000167349
|
si:dkey-283b1.6
|
si:dkey-283b1.6 |
| chr20_-_25533739 | 5.29 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr13_-_10945288 | 5.26 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr8_+_16004154 | 5.19 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr18_-_47533692 | 5.17 |
ENSDART00000191097
|
CABZ01112461.1
|
|
| chr8_-_49431939 | 5.00 |
ENSDART00000011453
ENSDART00000088240 ENSDART00000114173 |
sypb
|
synaptophysin b |
| chr16_-_12953739 | 4.99 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr25_+_7982979 | 4.92 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr3_+_16762483 | 4.87 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr12_-_20350629 | 4.73 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr20_-_25671342 | 4.69 |
ENSDART00000182775
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr23_+_20408227 | 4.65 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr8_+_24861264 | 4.57 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr14_-_18672561 | 4.53 |
ENSDART00000166730
ENSDART00000006998 |
slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr13_+_47821524 | 4.52 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr23_-_35483163 | 4.39 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr1_+_41666611 | 4.27 |
ENSDART00000145789
|
fbxo41
|
F-box protein 41 |
| chr23_+_25172682 | 4.24 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr19_+_37857936 | 4.20 |
ENSDART00000189289
|
nxph1
|
neurexophilin 1 |
| chr8_+_26268726 | 4.14 |
ENSDART00000180883
|
slc26a6
|
solute carrier family 26, member 6 |
| chr10_+_10738880 | 4.12 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr15_+_26600611 | 4.02 |
ENSDART00000155352
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr2_-_30770736 | 4.02 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr13_+_2908764 | 3.99 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr9_+_17971935 | 3.97 |
ENSDART00000149736
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr16_-_22775480 | 3.74 |
ENSDART00000141778
ENSDART00000145585 ENSDART00000125963 ENSDART00000127570 |
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr7_+_26138240 | 3.68 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr5_-_12524996 | 3.61 |
ENSDART00000142258
|
si:ch73-263f13.1
|
si:ch73-263f13.1 |
| chr20_+_25879826 | 3.60 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr21_+_12010505 | 3.57 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr13_-_20381485 | 3.57 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr24_-_11908115 | 3.56 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr11_-_1400507 | 3.52 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr15_+_7992906 | 3.47 |
ENSDART00000090790
|
cadm2b
|
cell adhesion molecule 2b |
| chr6_+_58915889 | 3.46 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr3_+_17251542 | 3.46 |
ENSDART00000024832
|
stat5a
|
signal transducer and activator of transcription 5a |
| chr18_-_14941840 | 3.44 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr10_-_43029001 | 3.38 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr7_+_54642005 | 3.32 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr24_+_29381946 | 3.28 |
ENSDART00000189551
|
ntng1a
|
netrin g1a |
| chr6_-_38419318 | 3.28 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr5_-_57879138 | 3.27 |
ENSDART00000145959
|
sik2a
|
salt-inducible kinase 2a |
| chr9_-_42871756 | 3.26 |
ENSDART00000191396
|
ttn.1
|
titin, tandem duplicate 1 |
| chr6_-_38418862 | 3.25 |
ENSDART00000104135
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr20_+_46897504 | 3.24 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr14_+_46268248 | 3.23 |
ENSDART00000172979
|
cabp2b
|
calcium binding protein 2b |
| chr5_-_12525441 | 3.21 |
ENSDART00000177490
ENSDART00000164190 |
ksr2
|
kinase suppressor of ras 2 |
| chr6_-_40697585 | 3.20 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr2_+_35240764 | 3.17 |
ENSDART00000015827
|
tnr
|
tenascin R (restrictin, janusin) |
| chr20_-_27390258 | 3.13 |
ENSDART00000010584
|
prph2l
|
peripherin 2, like |
| chr6_-_49078702 | 3.12 |
ENSDART00000135185
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr18_-_6803424 | 3.10 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr19_-_46777963 | 3.09 |
ENSDART00000046857
|
abrab
|
actin binding Rho activating protein b |
| chr17_+_39242437 | 3.08 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr2_+_33052360 | 3.03 |
ENSDART00000134651
|
rnf220a
|
ring finger protein 220a |
| chr14_+_35691889 | 3.01 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr9_+_29585943 | 2.93 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr4_+_26628822 | 2.89 |
ENSDART00000191030
ENSDART00000186113 ENSDART00000186764 ENSDART00000165158 |
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr22_+_15336752 | 2.88 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr14_+_17197132 | 2.86 |
ENSDART00000054598
|
rtn4rl2b
|
reticulon 4 receptor-like 2b |
| chr7_+_48460239 | 2.86 |
ENSDART00000052113
|
lingo1b
|
leucine rich repeat and Ig domain containing 1b |
| chr13_+_835390 | 2.85 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr18_+_23249519 | 2.81 |
ENSDART00000005740
ENSDART00000147446 ENSDART00000124818 |
mef2aa
|
myocyte enhancer factor 2aa |
| chr14_-_1313480 | 2.80 |
ENSDART00000097748
|
il21
|
interleukin 21 |
| chr15_-_568645 | 2.79 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr25_-_13728111 | 2.76 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr7_-_39203799 | 2.74 |
ENSDART00000173727
|
chrm4a
|
cholinergic receptor, muscarinic 4a |
| chr14_-_41556720 | 2.71 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr17_+_45654724 | 2.71 |
ENSDART00000098952
|
zgc:162184
|
zgc:162184 |
| chr5_-_42272517 | 2.68 |
ENSDART00000137692
ENSDART00000164363 |
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr24_-_39858710 | 2.67 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr23_+_27912079 | 2.65 |
ENSDART00000171859
|
BX539336.1
|
|
| chr25_+_4091067 | 2.63 |
ENSDART00000104926
|
eps8l2
|
EPS8 like 2 |
| chr2_+_38261748 | 2.55 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr22_-_17052381 | 2.52 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr24_+_29382109 | 2.50 |
ENSDART00000184620
ENSDART00000188414 ENSDART00000186132 ENSDART00000191489 |
ntng1a
|
netrin g1a |
| chr5_+_31049742 | 2.48 |
ENSDART00000173097
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr2_-_24289641 | 2.38 |
ENSDART00000128784
ENSDART00000123565 ENSDART00000141922 ENSDART00000184550 ENSDART00000191469 |
myh7l
|
myosin heavy chain 7-like |
| chr8_+_7778770 | 2.34 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr5_+_26728115 | 2.28 |
ENSDART00000098545
|
tmem150aa
|
transmembrane protein 150Aa |
| chr1_+_41854298 | 2.27 |
ENSDART00000192672
|
smox
|
spermine oxidase |
| chr12_+_23850661 | 2.24 |
ENSDART00000152921
|
svila
|
supervillin a |
| chr22_-_11626014 | 2.24 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr2_-_6482240 | 2.23 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr24_+_29449690 | 2.19 |
ENSDART00000105743
ENSDART00000193556 ENSDART00000145816 |
ntng1a
|
netrin g1a |
| chr6_-_49078263 | 2.18 |
ENSDART00000032982
|
slc5a8l
|
solute carrier family 5 (iodide transporter), member 8-like |
| chr13_+_844150 | 2.16 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr1_+_54137089 | 2.10 |
ENSDART00000062945
|
LO017798.1
|
|
| chr3_-_35602233 | 2.01 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr14_-_2322484 | 2.01 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr1_-_38816685 | 1.99 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr13_+_23677949 | 1.97 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr20_+_40150612 | 1.93 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr7_+_9904627 | 1.88 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr1_+_494873 | 1.87 |
ENSDART00000134581
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr19_+_7567763 | 1.82 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr15_-_2903475 | 1.82 |
ENSDART00000043226
|
guca1c
|
guanylate cyclase activator 1C |
| chr22_-_16400484 | 1.82 |
ENSDART00000135987
|
lama3
|
laminin, alpha 3 |
| chr2_-_52550135 | 1.81 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr8_+_25761654 | 1.79 |
ENSDART00000137899
ENSDART00000062403 |
tmem9
|
transmembrane protein 9 |
| chr25_+_16098620 | 1.78 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr19_-_7321221 | 1.70 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr18_+_45228691 | 1.66 |
ENSDART00000127953
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr8_+_6576940 | 1.65 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr10_+_25222367 | 1.62 |
ENSDART00000042767
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr25_+_33002963 | 1.62 |
ENSDART00000187366
|
CABZ01046980.1
|
|
| chr13_-_27388097 | 1.61 |
ENSDART00000187472
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr23_-_33750307 | 1.61 |
ENSDART00000162772
|
bin2a
|
bridging integrator 2a |
| chr2_-_37312927 | 1.59 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr12_-_13081229 | 1.59 |
ENSDART00000152510
ENSDART00000126866 |
ccser2b
|
coiled-coil serine-rich protein 2b |
| chr8_-_25846188 | 1.58 |
ENSDART00000128829
|
efhd2
|
EF-hand domain family, member D2 |
| chr7_-_50604626 | 1.58 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr5_-_16425781 | 1.57 |
ENSDART00000185624
ENSDART00000180617 |
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr13_-_4848889 | 1.57 |
ENSDART00000165259
|
mcu
|
mitochondrial calcium uniporter |
| chr23_-_33750135 | 1.56 |
ENSDART00000187641
|
bin2a
|
bridging integrator 2a |
| chr10_-_2527342 | 1.56 |
ENSDART00000184168
|
CU856539.1
|
|
| chr8_+_4838114 | 1.54 |
ENSDART00000146667
|
es1
|
es1 protein |
| chr11_-_6265574 | 1.53 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr20_-_7000225 | 1.52 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr5_-_37103487 | 1.50 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr25_-_23052707 | 1.50 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr5_-_24750286 | 1.49 |
ENSDART00000089968
|
rasl10a
|
RAS-like, family 10, member A |
| chr16_+_54829293 | 1.49 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr16_-_2504005 | 1.48 |
ENSDART00000089944
|
wdr26a
|
WD repeat domain 26a |
| chr8_+_16004551 | 1.46 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_46368520 | 1.46 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr6_+_51932267 | 1.45 |
ENSDART00000156256
|
angpt4
|
angiopoietin 4 |
| chr1_-_17570013 | 1.44 |
ENSDART00000146946
|
acsl1a
|
acyl-CoA synthetase long chain family member 1a |
| chr6_-_35488180 | 1.43 |
ENSDART00000183258
|
rgs8
|
regulator of G protein signaling 8 |
| chr2_-_52841762 | 1.39 |
ENSDART00000114682
|
ralbp1
|
ralA binding protein 1 |
| chr17_-_28770800 | 1.39 |
ENSDART00000156485
|
opn6b
|
opsin 6, group member b |
| chr15_-_21877726 | 1.31 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr5_+_19165949 | 1.31 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr20_-_40766387 | 1.30 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr23_+_9522781 | 1.29 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr10_+_10801564 | 1.27 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr6_-_13308813 | 1.27 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr2_-_38261272 | 1.27 |
ENSDART00000143743
|
si:ch211-14a17.10
|
si:ch211-14a17.10 |
| chr18_+_32615075 | 1.21 |
ENSDART00000166937
|
CABZ01012885.1
|
|
| chr17_+_19630272 | 1.14 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr8_-_14151051 | 1.13 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr17_+_23554932 | 1.10 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr5_+_29851433 | 1.05 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr9_-_45149695 | 1.05 |
ENSDART00000162033
|
CU467837.1
|
|
| chr5_-_13645995 | 1.04 |
ENSDART00000099665
ENSDART00000166957 |
purba
|
purine-rich element binding protein Ba |
| chr1_+_52563298 | 1.04 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr9_+_11379782 | 1.03 |
ENSDART00000190282
ENSDART00000007308 |
wnt10a
|
wingless-type MMTV integration site family, member 10a |
| chr22_-_36856405 | 1.03 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr5_-_66301142 | 1.03 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr20_+_18992679 | 1.02 |
ENSDART00000147105
ENSDART00000152811 |
tdh
|
L-threonine dehydrogenase |
| chr9_+_2522797 | 0.97 |
ENSDART00000186786
ENSDART00000147034 |
gpr155a
|
G protein-coupled receptor 155a |
| chr13_-_23031803 | 0.97 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr20_+_33875256 | 0.97 |
ENSDART00000002554
|
rxrgb
|
retinoid X receptor, gamma b |
| chr13_-_29406534 | 0.96 |
ENSDART00000100877
|
zgc:153142
|
zgc:153142 |
| chr6_-_33129045 | 0.95 |
ENSDART00000073757
|
tspan1
|
tetraspanin 1 |
| chr14_+_2487672 | 0.94 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr5_-_26247973 | 0.93 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr12_-_20665164 | 0.93 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr16_+_38820486 | 0.93 |
ENSDART00000131866
|
trhra
|
thyrotropin-releasing hormone receptor a |
| chr8_+_20994433 | 0.92 |
ENSDART00000141736
|
si:dkeyp-82a1.8
|
si:dkeyp-82a1.8 |
| chr5_+_23151169 | 0.92 |
ENSDART00000125638
|
tbx5b
|
T-box 5b |
| chr2_+_15776408 | 0.91 |
ENSDART00000176628
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr6_-_33129312 | 0.90 |
ENSDART00000156987
|
tspan1
|
tetraspanin 1 |
| chr24_+_10202718 | 0.89 |
ENSDART00000126668
|
pou6f2
|
POU class 6 homeobox 2 |
| chr17_-_26868169 | 0.89 |
ENSDART00000157204
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr13_+_22479988 | 0.89 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr18_+_18000695 | 0.88 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr8_-_1219815 | 0.86 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr4_+_18824959 | 0.86 |
ENSDART00000146141
ENSDART00000040424 |
slc26a3.1
|
solute carrier family 26 (anion exchanger), member 3 |
| chr13_+_22480496 | 0.84 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr7_-_68419830 | 0.81 |
ENSDART00000191606
|
CU468723.2
|
|
| chr1_+_25848231 | 0.80 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr19_-_40985308 | 0.80 |
ENSDART00000132986
ENSDART00000142360 |
si:ch211-120e1.7
|
si:ch211-120e1.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of gata1b+gata2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 2.1 | 6.2 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 2.0 | 6.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.7 | 10.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 1.5 | 15.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 1.5 | 4.6 | GO:0015824 | proline transport(GO:0015824) |
| 1.4 | 11.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.9 | 3.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.7 | 7.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.6 | 4.1 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.6 | 3.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.5 | 1.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.5 | 3.7 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.5 | 3.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.5 | 1.6 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.5 | 1.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.4 | 2.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.4 | 12.7 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.4 | 1.4 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 1.0 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.3 | 4.9 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.3 | 4.6 | GO:1905144 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.3 | 3.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.3 | 0.9 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.3 | 5.0 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.3 | 6.5 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.3 | 4.5 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 | 5.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 1.6 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.3 | 1.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 11.0 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.3 | 5.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.6 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.2 | 1.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 3.0 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.2 | 3.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 5.0 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.2 | 2.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 3.3 | GO:0031034 | myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.2 | 2.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.2 | 4.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 4.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.0 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 5.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 9.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.0 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 3.1 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.1 | 3.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 2.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 5.3 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 3.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 1.0 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.9 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 3.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.9 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.1 | 0.9 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.4 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 1.0 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 2.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 9.0 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.1 | 1.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.1 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 5.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.1 | 2.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 3.8 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 3.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.6 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 3.5 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.9 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 2.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 3.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.0 | 1.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 5.4 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 1.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 1.5 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.0 | 2.0 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 1.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.7 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 2.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.9 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 8.3 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.6 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 3.1 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 3.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 5.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 8.0 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 11.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 8.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 6.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 0.6 | GO:0043220 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.2 | 9.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 6.6 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 6.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 1.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 1.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 4.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 6.3 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 3.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 4.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 4.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 3.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.0 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 6.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 25.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 9.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 17.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.3 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 11.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 2.2 | 8.8 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 1.5 | 13.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.3 | 5.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 1.2 | 6.2 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 1.1 | 15.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.7 | 7.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.7 | 9.0 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.7 | 19.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.6 | 3.2 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.6 | 4.9 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.6 | 4.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.6 | 11.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 6.3 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 1.6 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.5 | 6.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.5 | 2.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.4 | 3.6 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.4 | 5.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 3.0 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 5.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 1.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 4.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.3 | 1.7 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.3 | 1.3 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.3 | 1.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 2.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 1.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.2 | 1.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 7.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 1.0 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 1.8 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.2 | 6.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 0.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 6.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.2 | 1.0 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 5.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.7 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.7 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 1.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 2.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 2.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 1.0 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 2.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 3.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 3.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.3 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 5.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 13.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 5.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 2.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.4 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 7.7 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 1.3 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 9.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 5.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 3.7 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.0 | 1.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 3.0 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 4.6 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 2.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 3.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.2 | 1.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 2.8 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 2.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 4.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 3.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 4.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 14.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.2 | 3.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.0 | 11.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 6.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 3.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 3.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 3.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.3 | 2.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 4.1 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 10.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 4.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |