Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
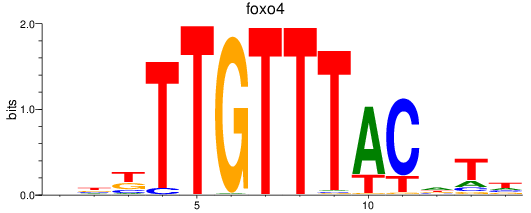
Results for foxo4
Z-value: 0.80
Transcription factors associated with foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo4
|
ENSDARG00000055792 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo4 | dr11_v1_chr14_+_30910114_30910114 | 0.79 | 5.7e-05 | Click! |
Activity profile of foxo4 motif
Sorted Z-values of foxo4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_24002503 | 1.86 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr13_-_33207367 | 1.71 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr15_+_46313082 | 1.69 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr3_-_60142530 | 1.46 |
ENSDART00000153247
|
si:ch211-120g10.1
|
si:ch211-120g10.1 |
| chr13_-_42400647 | 1.20 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr17_+_33158350 | 1.16 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr11_+_6819050 | 1.13 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr13_-_23665580 | 1.11 |
ENSDART00000144282
|
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr22_+_38276024 | 1.06 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr4_-_1720648 | 1.05 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr21_-_41588129 | 1.04 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr23_-_44677315 | 1.02 |
ENSDART00000143054
|
kif1c
|
kinesin family member 1C |
| chr18_+_41561285 | 1.02 |
ENSDART00000169621
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr5_+_36611128 | 0.94 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr2_-_26720854 | 0.92 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr15_+_30157602 | 0.90 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr2_-_31376606 | 0.89 |
ENSDART00000098988
ENSDART00000125746 |
clul1
|
clusterin-like 1 (retinal) |
| chr20_+_30445971 | 0.87 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr5_-_40190949 | 0.87 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr22_-_3449282 | 0.86 |
ENSDART00000136798
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr16_-_13612650 | 0.85 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr22_-_3914162 | 0.84 |
ENSDART00000187174
ENSDART00000190612 ENSDART00000187928 ENSDART00000057224 ENSDART00000184758 |
mhc1uma
|
major histocompatibility complex class I UMA |
| chr10_+_573667 | 0.84 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr2_-_50372467 | 0.83 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein like 2b |
| chr4_-_14926637 | 0.83 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr9_+_32358514 | 0.82 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr1_-_9940494 | 0.81 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr9_+_2343096 | 0.80 |
ENSDART00000062292
ENSDART00000191722 ENSDART00000135180 |
atf2
|
activating transcription factor 2 |
| chr22_+_3914318 | 0.78 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr15_+_46386261 | 0.77 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr3_+_57038033 | 0.77 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr17_+_44030692 | 0.76 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_-_13613475 | 0.75 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr7_+_38278860 | 0.75 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr3_-_60711127 | 0.75 |
ENSDART00000184119
|
ubald2
|
UBA-like domain containing 2 |
| chr5_+_51079504 | 0.73 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr4_+_18804317 | 0.71 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr19_-_7272921 | 0.71 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr25_+_22017182 | 0.71 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr9_+_17423941 | 0.70 |
ENSDART00000112884
ENSDART00000155233 |
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr17_-_7218481 | 0.70 |
ENSDART00000181967
|
SAMD5
|
sterile alpha motif domain containing 5 |
| chr10_-_36633882 | 0.69 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr10_-_33343244 | 0.69 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr21_-_2707768 | 0.68 |
ENSDART00000165384
|
CABZ01101739.1
|
|
| chr15_-_18574716 | 0.67 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr20_-_19365875 | 0.65 |
ENSDART00000063703
ENSDART00000187707 ENSDART00000161065 |
si:dkey-71h2.2
|
si:dkey-71h2.2 |
| chr22_+_26798853 | 0.65 |
ENSDART00000087576
ENSDART00000179780 |
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr21_-_13661631 | 0.65 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr9_+_9502610 | 0.64 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr1_+_51191049 | 0.64 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr5_+_55934129 | 0.63 |
ENSDART00000050969
|
tmem150ab
|
transmembrane protein 150Ab |
| chr2_+_33189582 | 0.62 |
ENSDART00000145588
ENSDART00000136330 ENSDART00000139295 ENSDART00000086340 |
rnf220a
|
ring finger protein 220a |
| chr9_+_24088062 | 0.62 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr3_-_12227359 | 0.61 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr2_-_24603325 | 0.61 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr20_-_25533739 | 0.60 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr6_+_48862 | 0.59 |
ENSDART00000082954
|
mbd5
|
methyl-CpG binding domain protein 5 |
| chr19_+_40115977 | 0.59 |
ENSDART00000139802
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr12_-_22509069 | 0.59 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr2_-_38035235 | 0.58 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr6_-_32999646 | 0.58 |
ENSDART00000159510
|
adcyap1r1b
|
adenylate cyclase activating polypeptide 1b (pituitary) receptor type I |
| chr8_+_16990120 | 0.57 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr15_+_30158652 | 0.57 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr14_-_33177935 | 0.57 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr23_-_7755373 | 0.57 |
ENSDART00000162868
|
PCMTD2 (1 of many)
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr15_-_30984557 | 0.57 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr23_-_12158685 | 0.57 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr5_-_68058168 | 0.56 |
ENSDART00000177026
|
rnf167
|
ring finger protein 167 |
| chr23_-_30787932 | 0.56 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr8_+_4803906 | 0.56 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr7_+_33372680 | 0.55 |
ENSDART00000193436
ENSDART00000099988 |
glceb
|
glucuronic acid epimerase b |
| chr19_+_20724347 | 0.55 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr5_+_44846434 | 0.55 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr12_-_30338779 | 0.55 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr2_-_57227542 | 0.54 |
ENSDART00000182675
ENSDART00000159480 |
btbd2b
|
BTB (POZ) domain containing 2b |
| chr2_-_43850637 | 0.54 |
ENSDART00000136077
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr5_+_62340799 | 0.53 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr24_-_5932982 | 0.53 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr5_+_1965296 | 0.53 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr6_-_35309661 | 0.52 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr19_+_2275019 | 0.52 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr17_+_24632440 | 0.51 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr7_+_9904627 | 0.51 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr13_-_43599898 | 0.51 |
ENSDART00000084416
ENSDART00000145705 |
ablim1a
|
actin binding LIM protein 1a |
| chr5_-_18046053 | 0.51 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr25_+_34938317 | 0.51 |
ENSDART00000042678
|
vps4a
|
vacuolar protein sorting 4a homolog A (S. cerevisiae) |
| chr22_-_28777374 | 0.50 |
ENSDART00000188206
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr18_+_26829086 | 0.49 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr20_-_16849306 | 0.49 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr3_+_24458899 | 0.49 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr2_-_52841762 | 0.49 |
ENSDART00000114682
|
ralbp1
|
ralA binding protein 1 |
| chr23_+_10469955 | 0.48 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr1_-_23596391 | 0.48 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr18_+_26829362 | 0.47 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr18_-_25177230 | 0.47 |
ENSDART00000013363
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr6_-_10305918 | 0.47 |
ENSDART00000090994
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr18_+_41560822 | 0.46 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr22_-_22231720 | 0.45 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr17_-_43399896 | 0.45 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr22_-_18179214 | 0.45 |
ENSDART00000129576
|
si:ch211-125m10.6
|
si:ch211-125m10.6 |
| chr8_+_17869225 | 0.45 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr18_-_21047580 | 0.44 |
ENSDART00000010189
|
igf1ra
|
insulin-like growth factor 1a receptor |
| chr10_+_213878 | 0.44 |
ENSDART00000135903
ENSDART00000138812 |
mpzl1l
|
myelin protein zero-like 1 like |
| chr11_+_39959039 | 0.43 |
ENSDART00000024304
|
per3
|
period circadian clock 3 |
| chr7_+_40205394 | 0.43 |
ENSDART00000173742
ENSDART00000148390 |
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr11_+_2710530 | 0.43 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr2_+_55365727 | 0.43 |
ENSDART00000162943
|
FP245456.1
|
|
| chr13_+_1100197 | 0.42 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr23_-_1571682 | 0.42 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr21_-_13662237 | 0.41 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr20_+_4060839 | 0.41 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr2_-_44971551 | 0.41 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr17_+_25856671 | 0.41 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr8_+_8643901 | 0.40 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr20_-_2949028 | 0.40 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr12_+_27213733 | 0.39 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr7_-_50604626 | 0.39 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr16_+_22587661 | 0.39 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr19_+_22074468 | 0.38 |
ENSDART00000136294
ENSDART00000090476 |
atp9b
|
ATPase phospholipid transporting 9B |
| chr17_-_2690083 | 0.38 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr5_-_18897482 | 0.38 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr10_+_28428222 | 0.38 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr18_-_14836862 | 0.38 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr18_-_16590056 | 0.38 |
ENSDART00000143744
|
mgat4c
|
mgat4 family, member C |
| chr20_-_40119872 | 0.37 |
ENSDART00000191569
|
nkain2
|
sodium/potassium transporting ATPase interacting 2 |
| chr16_-_26820634 | 0.37 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr9_+_41459759 | 0.37 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr21_-_42202792 | 0.37 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr14_+_8710568 | 0.37 |
ENSDART00000169505
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr5_+_22836364 | 0.37 |
ENSDART00000131885
|
si:ch211-26b3.2
|
si:ch211-26b3.2 |
| chr10_-_41156348 | 0.37 |
ENSDART00000058622
|
aak1b
|
AP2 associated kinase 1b |
| chr4_+_15954293 | 0.37 |
ENSDART00000132695
|
si:dkey-117n7.4
|
si:dkey-117n7.4 |
| chr13_+_28701233 | 0.37 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr8_+_17838924 | 0.36 |
ENSDART00000170762
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr25_-_17528994 | 0.36 |
ENSDART00000061712
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr25_-_4525081 | 0.36 |
ENSDART00000184347
|
pidd1
|
p53-induced death domain protein 1 |
| chr15_+_39977461 | 0.36 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr2_-_5942115 | 0.36 |
ENSDART00000154489
|
tmem125b
|
transmembrane protein 125b |
| chr11_-_25733910 | 0.36 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr16_-_22251414 | 0.35 |
ENSDART00000158500
ENSDART00000179998 |
atp8b2
|
ATPase phospholipid transporting 8B2 |
| chr3_-_51912019 | 0.35 |
ENSDART00000149914
|
aatka
|
apoptosis-associated tyrosine kinase a |
| chr12_+_27704015 | 0.35 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr15_+_20543770 | 0.35 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr5_-_50371205 | 0.35 |
ENSDART00000157800
|
si:ch73-280o22.2
|
si:ch73-280o22.2 |
| chr23_+_17981127 | 0.34 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr19_+_12237945 | 0.34 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr2_-_58142854 | 0.34 |
ENSDART00000169909
|
CABZ01110881.1
|
|
| chr24_+_14240196 | 0.34 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr12_-_979789 | 0.33 |
ENSDART00000128188
|
daglb
|
diacylglycerol lipase, beta |
| chr25_-_3139805 | 0.33 |
ENSDART00000166625
|
epx
|
eosinophil peroxidase |
| chr5_-_20678300 | 0.33 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr6_-_30839763 | 0.33 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr21_-_9782502 | 0.33 |
ENSDART00000158836
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr3_-_45298487 | 0.33 |
ENSDART00000102245
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr15_+_20843161 | 0.33 |
ENSDART00000153726
|
ulk2
|
unc-51 like autophagy activating kinase 2 |
| chr19_-_27315062 | 0.33 |
ENSDART00000141203
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr6_-_43882696 | 0.33 |
ENSDART00000064938
|
foxp1b
|
forkhead box P1b |
| chr24_-_19718077 | 0.32 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr11_+_21586335 | 0.32 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr14_-_25452503 | 0.32 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr17_+_19730001 | 0.32 |
ENSDART00000155025
|
grem2a
|
gremlin 2, DAN family BMP antagonist a |
| chr22_+_4707663 | 0.32 |
ENSDART00000042194
|
cers4a
|
ceramide synthase 4a |
| chr1_+_45839927 | 0.32 |
ENSDART00000148086
ENSDART00000180413 ENSDART00000048191 ENSDART00000179047 |
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr5_-_4931266 | 0.31 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr23_-_27608257 | 0.31 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr25_-_3647277 | 0.31 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr6_-_9565526 | 0.31 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr2_-_42492201 | 0.31 |
ENSDART00000180762
ENSDART00000009093 |
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr15_-_44052927 | 0.31 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr13_-_23264724 | 0.30 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr1_-_37383741 | 0.30 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr22_+_1868983 | 0.30 |
ENSDART00000160135
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr22_+_23546926 | 0.30 |
ENSDART00000157940
|
aspm
|
abnormal spindle microtubule assembly |
| chr15_-_25365570 | 0.30 |
ENSDART00000152754
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr6_+_7466223 | 0.30 |
ENSDART00000148908
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr2_-_12243213 | 0.30 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr16_+_42667560 | 0.30 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr16_-_5612480 | 0.30 |
ENSDART00000157944
|
CR848788.1
|
|
| chr22_+_2660505 | 0.29 |
ENSDART00000135555
ENSDART00000031485 |
znf1162
|
zinc finger protein 1162 |
| chr4_-_14926106 | 0.29 |
ENSDART00000147629
|
prdm4
|
PR domain containing 4 |
| chr7_+_13995792 | 0.29 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr24_+_21192431 | 0.29 |
ENSDART00000152027
|
usf3
|
upstream transcription factor family member 3 |
| chr13_-_4333181 | 0.29 |
ENSDART00000122406
|
znf318
|
zinc finger protein 318 |
| chr24_-_23323526 | 0.29 |
ENSDART00000112256
ENSDART00000176903 |
zfhx4
|
zinc finger homeobox 4 |
| chr16_+_38337783 | 0.29 |
ENSDART00000135008
|
gabpb2b
|
GA binding protein transcription factor, beta subunit 2b |
| chr7_-_51749683 | 0.28 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr15_-_5720583 | 0.28 |
ENSDART00000158034
ENSDART00000190332 ENSDART00000109599 |
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr9_-_10532591 | 0.28 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr10_-_36214582 | 0.28 |
ENSDART00000166471
|
or109-11
|
odorant receptor, family D, subfamily 109, member 11 |
| chr21_+_40276569 | 0.28 |
ENSDART00000144436
|
si:ch211-218m3.13
|
si:ch211-218m3.13 |
| chr8_+_40081403 | 0.28 |
ENSDART00000138036
|
lrrc75ba
|
leucine rich repeat containing 75Ba |
| chr13_+_9368621 | 0.28 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr20_-_40119090 | 0.28 |
ENSDART00000145592
|
nkain2
|
sodium/potassium transporting ATPase interacting 2 |
| chr20_-_52928541 | 0.28 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr14_-_36378494 | 0.28 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr7_+_60551133 | 0.27 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr15_-_3976035 | 0.27 |
ENSDART00000168061
|
si:ch73-309g22.1
|
si:ch73-309g22.1 |
| chr9_-_2572790 | 0.27 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr3_+_55031685 | 0.27 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr18_-_14836600 | 0.27 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 0.8 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.2 | 2.2 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 0.8 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.2 | 0.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 0.5 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.8 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 1.0 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.8 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.5 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.6 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.4 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.8 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.3 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.2 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.1 | 0.3 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 1.4 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.0 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.3 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.1 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.9 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.3 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.0 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.6 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.8 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.3 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.4 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.7 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 1.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.0 | 0.4 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.8 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.1 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0086009 | membrane repolarization(GO:0086009) |
| 0.0 | 0.0 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.8 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 1.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 2.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0052726 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.6 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 1.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 0.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.5 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.4 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.0 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 1.0 | GO:0015370 | solute:sodium symporter activity(GO:0015370) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.4 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.1 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 1.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |