Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
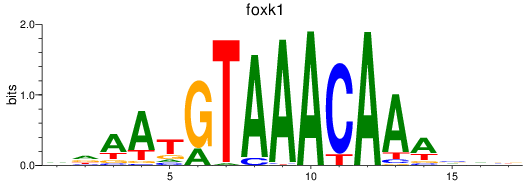
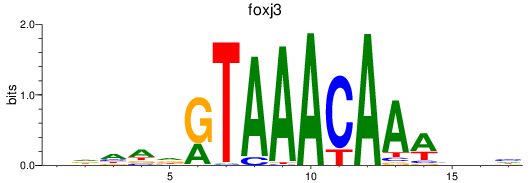
Results for foxk1_foxj3
Z-value: 0.38
Transcription factors associated with foxk1_foxj3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxk1
|
ENSDARG00000037872 | forkhead box K1 |
|
foxj3
|
ENSDARG00000075774 | forkhead box J3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxk1 | dr11_v1_chr3_-_40933415_40933415 | 0.15 | 5.3e-01 | Click! |
| foxj3 | dr11_v1_chr11_-_26362294_26362334 | -0.07 | 7.9e-01 | Click! |
Activity profile of foxk1_foxj3 motif
Sorted Z-values of foxk1_foxj3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_24305877 | 1.29 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr14_-_41478265 | 1.04 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr17_+_21295132 | 1.02 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr14_-_35462148 | 1.01 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr7_-_60831082 | 0.99 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr14_+_26439227 | 0.98 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr15_+_19990068 | 0.91 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr23_-_28025943 | 0.90 |
ENSDART00000181146
|
sp5l
|
Sp5 transcription factor-like |
| chr5_-_51998708 | 0.85 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr18_-_24988645 | 0.84 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr25_-_13408760 | 0.80 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr8_-_16697912 | 0.79 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr20_+_25904199 | 0.73 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr9_+_41459759 | 0.63 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr5_+_12528693 | 0.62 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr23_+_7692042 | 0.62 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr15_-_17960228 | 0.61 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr15_+_26933196 | 0.61 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr15_+_34069746 | 0.60 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr12_+_45238292 | 0.59 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr18_+_6857071 | 0.56 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr7_+_46019780 | 0.56 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr5_-_54672763 | 0.55 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr6_-_40922971 | 0.54 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr13_+_5978809 | 0.54 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr19_+_25971000 | 0.53 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr9_-_12885201 | 0.50 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr10_+_20070178 | 0.49 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr24_-_6678640 | 0.48 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr11_+_2710530 | 0.47 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr14_-_38843690 | 0.46 |
ENSDART00000183629
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr15_-_28805493 | 0.45 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr16_+_9580699 | 0.45 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_-_20446082 | 0.44 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr2_-_42109575 | 0.43 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr2_+_16696052 | 0.43 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr1_-_12394048 | 0.43 |
ENSDART00000146067
ENSDART00000134708 |
sclt1
|
sodium channel and clathrin linker 1 |
| chr5_+_26913120 | 0.42 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr23_-_17450746 | 0.41 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr5_+_22836364 | 0.41 |
ENSDART00000131885
|
si:ch211-26b3.2
|
si:ch211-26b3.2 |
| chr11_-_25257045 | 0.40 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr12_+_8822717 | 0.39 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr16_+_28578352 | 0.39 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr5_-_66749535 | 0.39 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr2_+_27386617 | 0.39 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr8_+_10305400 | 0.38 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr15_-_44052927 | 0.38 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr13_-_33207367 | 0.37 |
ENSDART00000146138
ENSDART00000109667 ENSDART00000182741 |
trip11
|
thyroid hormone receptor interactor 11 |
| chr18_+_25225524 | 0.37 |
ENSDART00000055563
|
si:dkeyp-59c12.1
|
si:dkeyp-59c12.1 |
| chr17_-_2690083 | 0.37 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr3_+_42999844 | 0.37 |
ENSDART00000183950
|
ints1
|
integrator complex subunit 1 |
| chr8_-_39859688 | 0.36 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr2_+_55365727 | 0.35 |
ENSDART00000162943
|
FP245456.1
|
|
| chr7_+_40205394 | 0.35 |
ENSDART00000173742
ENSDART00000148390 |
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr22_+_37631234 | 0.35 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr22_-_34979139 | 0.35 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr20_-_9428021 | 0.35 |
ENSDART00000025330
|
rdh14b
|
retinol dehydrogenase 14b |
| chr1_-_23370395 | 0.35 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr11_-_25257595 | 0.35 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr9_+_24088062 | 0.34 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr5_+_49744713 | 0.34 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr17_-_14966384 | 0.34 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr17_+_37215820 | 0.34 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr13_+_35635672 | 0.34 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr1_+_17593392 | 0.33 |
ENSDART00000078889
|
helt
|
helt bHLH transcription factor |
| chr10_+_39084354 | 0.33 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr3_+_42999277 | 0.33 |
ENSDART00000163595
|
ints1
|
integrator complex subunit 1 |
| chr16_-_26820634 | 0.33 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr11_+_18612421 | 0.33 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr5_+_13549603 | 0.32 |
ENSDART00000171270
|
ckap2l
|
cytoskeleton associated protein 2-like |
| chr23_-_3758637 | 0.31 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr4_-_4256300 | 0.31 |
ENSDART00000103319
ENSDART00000150279 |
cd9b
|
CD9 molecule b |
| chr6_-_8466717 | 0.31 |
ENSDART00000151577
ENSDART00000151800 ENSDART00000151227 |
si:dkey-217d24.6
|
si:dkey-217d24.6 |
| chr7_+_5530154 | 0.31 |
ENSDART00000181610
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr21_-_13661631 | 0.31 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr24_+_19542323 | 0.30 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr7_-_13906409 | 0.30 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr9_-_1990323 | 0.30 |
ENSDART00000082332
|
hoxd13a
|
homeobox D13a |
| chr13_-_23665580 | 0.30 |
ENSDART00000144282
|
map3k21
|
mitogen-activated protein kinase kinase kinase 21 |
| chr20_-_23171430 | 0.30 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr23_-_32304650 | 0.30 |
ENSDART00000143772
ENSDART00000085642 ENSDART00000188989 |
dgkaa
|
diacylglycerol kinase, alpha a |
| chr9_+_52411530 | 0.30 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr17_+_4325693 | 0.30 |
ENSDART00000154264
|
mcm8
|
minichromosome maintenance 8 homologous recombination repair factor |
| chr19_+_20177887 | 0.30 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr9_+_21306902 | 0.30 |
ENSDART00000138554
ENSDART00000004108 |
xpo4
|
exportin 4 |
| chr13_-_5257303 | 0.29 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr11_+_16153207 | 0.29 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr23_+_32029304 | 0.28 |
ENSDART00000185217
|
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr15_-_18232712 | 0.28 |
ENSDART00000081199
|
wu:fj20b03
|
wu:fj20b03 |
| chr12_+_19188542 | 0.28 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr19_-_22843480 | 0.28 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr11_+_16152316 | 0.28 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr16_+_26732086 | 0.27 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr24_+_37640626 | 0.27 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr6_-_39700965 | 0.27 |
ENSDART00000156645
|
espl1
|
extra spindle pole bodies like 1, separase |
| chr8_-_1267247 | 0.27 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr8_+_26034623 | 0.27 |
ENSDART00000004521
ENSDART00000142555 |
arih2
|
ariadne homolog 2 (Drosophila) |
| chr17_+_24718272 | 0.27 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr13_+_29925397 | 0.27 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr7_-_46019756 | 0.27 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr23_-_10722664 | 0.27 |
ENSDART00000146526
ENSDART00000129022 ENSDART00000104985 |
foxp1a
|
forkhead box P1a |
| chr8_-_16712111 | 0.27 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr11_+_25257022 | 0.26 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr14_+_94946 | 0.26 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr21_+_25071805 | 0.26 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr13_+_42602406 | 0.26 |
ENSDART00000133388
ENSDART00000147996 |
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr16_+_25011994 | 0.25 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr25_+_13662606 | 0.25 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr1_+_53945934 | 0.25 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr20_-_16849306 | 0.25 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr21_-_27272657 | 0.25 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr2_-_30200206 | 0.25 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr4_-_20135919 | 0.25 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr14_+_6962271 | 0.24 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr21_+_17051478 | 0.24 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr6_-_35309661 | 0.24 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr11_-_16152400 | 0.24 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr4_-_20135406 | 0.24 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr17_+_24036791 | 0.24 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr12_+_30726425 | 0.24 |
ENSDART00000153275
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr17_-_27266053 | 0.23 |
ENSDART00000110903
|
E2F2
|
si:ch211-160f23.5 |
| chr25_+_18953575 | 0.23 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr15_+_37589698 | 0.23 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr4_+_27100531 | 0.23 |
ENSDART00000115200
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr17_+_16755287 | 0.23 |
ENSDART00000080129
|
ston2
|
stonin 2 |
| chr5_-_24008997 | 0.23 |
ENSDART00000066645
|
eif1axa
|
eukaryotic translation initiation factor 1A, X-linked, a |
| chr16_-_41131578 | 0.23 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr22_-_16154771 | 0.22 |
ENSDART00000009464
|
slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr23_+_32028574 | 0.22 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr5_+_44846434 | 0.22 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr6_-_14038804 | 0.22 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr13_-_25842074 | 0.22 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr23_+_31596441 | 0.22 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr4_-_6373735 | 0.21 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr10_-_36633882 | 0.21 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr25_+_2263857 | 0.20 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr23_-_10723009 | 0.20 |
ENSDART00000189721
|
foxp1a
|
forkhead box P1a |
| chr19_-_47997424 | 0.20 |
ENSDART00000081675
|
ctnnb2
|
catenin, beta 2 |
| chr5_-_19014589 | 0.20 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr9_-_30555725 | 0.19 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr8_-_7567815 | 0.19 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr19_-_10881141 | 0.19 |
ENSDART00000162793
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr24_-_19718077 | 0.19 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr21_-_27273147 | 0.19 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr18_-_5875433 | 0.19 |
ENSDART00000151727
|
nob1
|
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr2_+_26179096 | 0.19 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr23_+_21978816 | 0.19 |
ENSDART00000087110
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr23_+_28077953 | 0.19 |
ENSDART00000186122
ENSDART00000111570 |
slc26a10
|
solute carrier family 26, member 10 |
| chr23_+_21978584 | 0.18 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr21_+_20386865 | 0.18 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr9_+_38163876 | 0.18 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr21_+_19547806 | 0.18 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr25_+_18954189 | 0.18 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr7_+_19851422 | 0.18 |
ENSDART00000142970
ENSDART00000190027 |
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr16_-_41714988 | 0.18 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr17_+_44030692 | 0.18 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr16_-_30847192 | 0.17 |
ENSDART00000191106
ENSDART00000128158 |
ptk2ab
|
protein tyrosine kinase 2ab |
| chr10_+_31809226 | 0.17 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr2_+_19195841 | 0.17 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr20_+_53368611 | 0.17 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr13_+_9368621 | 0.16 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr15_-_7337537 | 0.16 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr23_+_25354856 | 0.16 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr5_-_16351306 | 0.16 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr6_-_43792179 | 0.16 |
ENSDART00000160849
|
foxp1b
|
forkhead box P1b |
| chr23_-_45398622 | 0.16 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr1_+_35494837 | 0.16 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr5_+_53009083 | 0.16 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr2_+_21000334 | 0.16 |
ENSDART00000062563
ENSDART00000147809 |
rreb1b
|
ras responsive element binding protein 1b |
| chr14_+_35424539 | 0.16 |
ENSDART00000171809
ENSDART00000162185 |
sytl4
|
synaptotagmin-like 4 |
| chr15_-_4011013 | 0.16 |
ENSDART00000158029
|
CR626884.3
|
|
| chr12_-_5998898 | 0.16 |
ENSDART00000142659
ENSDART00000004896 |
kat7b
|
K(lysine) acetyltransferase 7b |
| chr18_+_5875268 | 0.15 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr9_+_9502610 | 0.15 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr10_-_17222083 | 0.15 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr16_+_28578648 | 0.15 |
ENSDART00000149566
|
nmt2
|
N-myristoyltransferase 2 |
| chr24_-_31306724 | 0.15 |
ENSDART00000165399
|
acp5b
|
acid phosphatase 5b, tartrate resistant |
| chr16_+_38350704 | 0.15 |
ENSDART00000140761
|
elp6
|
elongator acetyltransferase complex subunit 6 |
| chr1_+_51191049 | 0.15 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr2_-_26720854 | 0.15 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr11_-_21586157 | 0.15 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr22_+_24446964 | 0.15 |
ENSDART00000166812
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr2_-_23677422 | 0.15 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr7_-_51102479 | 0.15 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr2_+_38373272 | 0.15 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr10_-_17221725 | 0.14 |
ENSDART00000111088
|
depdc5
|
DEP domain containing 5 |
| chr16_-_49646625 | 0.14 |
ENSDART00000101629
|
efhb
|
EF-hand domain family, member B |
| chr24_-_37640705 | 0.14 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr21_-_26028205 | 0.14 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr23_-_27608257 | 0.14 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr23_+_31596194 | 0.14 |
ENSDART00000160748
|
tbpl1
|
TBP-like 1 |
| chr17_-_27273296 | 0.14 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr23_-_43424510 | 0.13 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr25_-_25142387 | 0.13 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr15_-_37589600 | 0.13 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr2_+_49572059 | 0.13 |
ENSDART00000108861
|
sema4e
|
semaphorin 4e |
| chr5_+_28271412 | 0.13 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr9_+_50600355 | 0.13 |
ENSDART00000187567
|
fign
|
fidgetin |
| chr13_+_29926094 | 0.12 |
ENSDART00000057528
|
cuedc2
|
CUE domain containing 2 |
| chr10_-_14929630 | 0.12 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxk1_foxj3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0016123 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.2 | 0.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.5 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 1.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.8 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.3 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.3 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.5 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.9 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.5 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.5 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.4 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 0.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.1 | GO:0036314 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 0.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.0 | 0.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.3 | GO:0032392 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.6 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.3 | 1.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.2 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.3 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.5 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.6 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.6 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 1.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |