Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
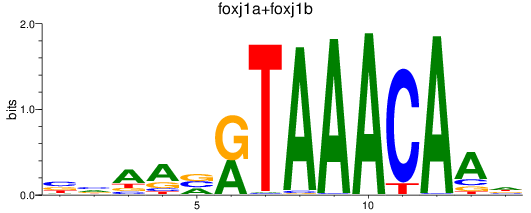
Results for foxj1a+foxj1b
Z-value: 1.57
Transcription factors associated with foxj1a+foxj1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj1b
|
ENSDARG00000088290 | forkhead box J1b |
|
foxj1a
|
ENSDARG00000101919 | forkhead box J1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj1a | dr11_v1_chr3_+_60721342_60721342 | 0.78 | 7.1e-05 | Click! |
| foxj1b | dr11_v1_chr12_+_20149707_20149707 | 0.58 | 1.0e-02 | Click! |
Activity profile of foxj1a+foxj1b motif
Sorted Z-values of foxj1a+foxj1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_20167206 | 4.83 |
ENSDART00000104874
ENSDART00000191995 |
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr14_-_41478265 | 4.62 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr10_-_35257458 | 4.18 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr8_-_16697912 | 3.57 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr14_-_35462148 | 3.54 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr15_+_26933196 | 3.54 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr16_+_40576679 | 3.49 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr1_+_9153141 | 3.40 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr15_+_34069746 | 3.31 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr2_+_19195841 | 3.24 |
ENSDART00000163137
ENSDART00000161095 |
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr7_-_28611145 | 3.14 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr17_-_28100741 | 3.12 |
ENSDART00000180532
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr14_+_26439227 | 3.12 |
ENSDART00000054183
|
gpr137
|
G protein-coupled receptor 137 |
| chr17_-_28100501 | 2.86 |
ENSDART00000149543
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr4_+_27100531 | 2.84 |
ENSDART00000115200
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr23_+_28077953 | 2.82 |
ENSDART00000186122
ENSDART00000111570 |
slc26a10
|
solute carrier family 26, member 10 |
| chr10_-_3416258 | 2.72 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr2_-_42958113 | 2.71 |
ENSDART00000139945
|
oc90
|
otoconin 90 |
| chr2_+_20866898 | 2.70 |
ENSDART00000150086
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr16_-_31435020 | 2.62 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr17_-_35881841 | 2.62 |
ENSDART00000110040
|
sox11a
|
SRY (sex determining region Y)-box 11a |
| chr7_-_33023404 | 2.59 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr9_-_1990323 | 2.55 |
ENSDART00000082332
|
hoxd13a
|
homeobox D13a |
| chr5_-_20446082 | 2.55 |
ENSDART00000051607
|
ISCU (1 of many)
|
si:ch211-191d15.2 |
| chr10_+_10728870 | 2.51 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr13_-_21650404 | 2.47 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr11_+_16153207 | 2.45 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr24_+_39211288 | 2.45 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr15_-_17960228 | 2.40 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr8_-_1267247 | 2.39 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr9_+_2041535 | 2.37 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr6_-_9695294 | 2.36 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr7_-_60831082 | 2.36 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr7_-_26270014 | 2.35 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr5_+_60847823 | 2.34 |
ENSDART00000074426
|
lig3
|
ligase III, DNA, ATP-dependent |
| chr13_+_18533005 | 2.34 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr20_+_15552657 | 2.27 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr4_-_6373735 | 2.26 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr15_-_44052927 | 2.24 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr13_+_5978809 | 2.23 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr9_+_41459759 | 2.23 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr13_-_31687925 | 2.23 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr18_+_6857071 | 2.22 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr10_+_22775253 | 2.21 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr19_+_20177887 | 2.21 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr25_-_31423493 | 2.20 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr12_+_18899396 | 2.16 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr6_-_14038804 | 2.14 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr7_+_38808027 | 2.13 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr24_+_19542323 | 2.12 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr13_-_31647323 | 2.12 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr2_+_38271392 | 2.10 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr7_-_50367642 | 2.08 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr16_+_26732086 | 2.08 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr24_-_25461267 | 2.06 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr18_+_20226843 | 2.06 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr1_+_53945934 | 2.05 |
ENSDART00000052838
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr10_-_23099809 | 2.04 |
ENSDART00000148333
ENSDART00000079703 ENSDART00000162444 |
nle1
|
notchless homolog 1 (Drosophila) |
| chr12_+_3912544 | 2.03 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr17_-_27273296 | 2.03 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr19_+_25971000 | 2.00 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr5_+_12528693 | 1.98 |
ENSDART00000051670
|
rfc5
|
replication factor C (activator 1) 5 |
| chr6_-_40922971 | 1.97 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr13_-_25842074 | 1.95 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr8_-_39859688 | 1.94 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr2_+_16696052 | 1.94 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr22_+_10232527 | 1.94 |
ENSDART00000139297
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr6_-_10728057 | 1.92 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr11_-_16152400 | 1.89 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr22_+_37631234 | 1.89 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr2_-_30200206 | 1.89 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr23_-_17450746 | 1.88 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr23_+_7692042 | 1.87 |
ENSDART00000018512
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr20_-_36800002 | 1.87 |
ENSDART00000015190
|
ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr11_+_16152316 | 1.83 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr19_-_22843480 | 1.83 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr18_-_24988645 | 1.79 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr25_+_19734038 | 1.79 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr16_-_41131578 | 1.79 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr7_-_26844064 | 1.78 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr16_-_7228276 | 1.77 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr17_+_24718272 | 1.77 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr23_-_43424510 | 1.76 |
ENSDART00000055564
|
rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr16_-_24605969 | 1.76 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr4_-_20135919 | 1.75 |
ENSDART00000172230
|
cep83
|
centrosomal protein 83 |
| chr16_+_33121260 | 1.73 |
ENSDART00000058472
|
akirin1
|
akirin 1 |
| chr15_-_25527580 | 1.71 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr13_+_29926094 | 1.71 |
ENSDART00000057528
|
cuedc2
|
CUE domain containing 2 |
| chr1_+_11107688 | 1.70 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr13_+_29773153 | 1.68 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr19_+_4968947 | 1.68 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr10_+_35257651 | 1.65 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr23_+_31596441 | 1.65 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr7_-_28549361 | 1.64 |
ENSDART00000173918
ENSDART00000054368 ENSDART00000113313 |
st5
|
suppression of tumorigenicity 5 |
| chr1_+_47178529 | 1.64 |
ENSDART00000158432
ENSDART00000074450 ENSDART00000137448 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr16_-_13595027 | 1.62 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr16_+_33121106 | 1.62 |
ENSDART00000110195
|
akirin1
|
akirin 1 |
| chr1_-_51720633 | 1.62 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr7_+_48805534 | 1.61 |
ENSDART00000145375
ENSDART00000148744 |
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr16_+_9580699 | 1.60 |
ENSDART00000165565
|
taf2
|
TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_+_23784842 | 1.60 |
ENSDART00000192889
ENSDART00000138375 |
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr3_-_36419641 | 1.58 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr2_+_1988036 | 1.58 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr6_-_10728921 | 1.57 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr13_+_29925397 | 1.57 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr7_+_48805725 | 1.56 |
ENSDART00000166543
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr6_-_37745508 | 1.55 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr19_+_5315987 | 1.55 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr4_-_12795030 | 1.53 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr5_-_19014589 | 1.53 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr15_-_28805493 | 1.52 |
ENSDART00000179617
|
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr11_+_18612421 | 1.52 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr11_+_14622379 | 1.51 |
ENSDART00000112589
|
efna2b
|
ephrin-A2b |
| chr6_+_43015916 | 1.51 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr8_+_49065348 | 1.51 |
ENSDART00000032277
|
ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr2_+_26179096 | 1.50 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr8_-_16712111 | 1.50 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr17_-_21793113 | 1.50 |
ENSDART00000104612
|
hmx3a
|
H6 family homeobox 3a |
| chr25_-_10571078 | 1.49 |
ENSDART00000153898
|
si:ch211-107e6.5
|
si:ch211-107e6.5 |
| chr24_+_9298198 | 1.49 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr5_-_24231139 | 1.49 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr16_-_21692024 | 1.49 |
ENSDART00000123597
|
si:ch211-154o6.2
|
si:ch211-154o6.2 |
| chr20_-_2667902 | 1.47 |
ENSDART00000036373
|
cfap206
|
cilia and flagella associated protein 206 |
| chr5_-_66749535 | 1.45 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr15_-_7337537 | 1.45 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr5_+_29726428 | 1.44 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr15_+_19990068 | 1.43 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr1_+_51537250 | 1.43 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr25_-_34740627 | 1.43 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr10_-_35186310 | 1.43 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr10_-_42237304 | 1.43 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr7_-_24112484 | 1.42 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr11_-_45420212 | 1.40 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr19_+_18739085 | 1.39 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr19_-_25693728 | 1.38 |
ENSDART00000136187
|
bloc1s4
|
biogenesis of lysosomal organelles complex-1, subunit 4, cappuccino |
| chr7_+_26844261 | 1.37 |
ENSDART00000079165
|
ext2
|
exostosin glycosyltransferase 2 |
| chr8_+_10305400 | 1.36 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr25_-_25142387 | 1.36 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr7_-_37555208 | 1.35 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr14_-_33425170 | 1.33 |
ENSDART00000124629
ENSDART00000105800 ENSDART00000001318 |
nkap
|
NFKB activating protein |
| chr21_+_25071805 | 1.33 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr16_-_41714988 | 1.32 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr1_+_45969240 | 1.32 |
ENSDART00000042086
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr14_-_34044369 | 1.30 |
ENSDART00000149396
ENSDART00000123607 ENSDART00000190746 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr19_-_46018152 | 1.29 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr17_+_16755287 | 1.29 |
ENSDART00000080129
|
ston2
|
stonin 2 |
| chr5_-_54672763 | 1.28 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr21_-_28920245 | 1.28 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr22_+_11153590 | 1.26 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr4_+_5868034 | 1.24 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr14_+_33264303 | 1.24 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr10_+_28428222 | 1.23 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr12_-_8070969 | 1.23 |
ENSDART00000020995
|
tmem26b
|
transmembrane protein 26b |
| chr18_-_45761868 | 1.22 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr7_-_50367326 | 1.21 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr12_+_28117365 | 1.21 |
ENSDART00000066290
|
UTS2R
|
urotensin 2 receptor |
| chr3_-_20793655 | 1.21 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr24_+_37640626 | 1.20 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr7_+_52761841 | 1.19 |
ENSDART00000111444
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr20_-_2641233 | 1.17 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr1_+_34295925 | 1.17 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr21_-_30031396 | 1.17 |
ENSDART00000157167
|
pwwp2a
|
PWWP domain containing 2A |
| chr23_+_43424518 | 1.17 |
ENSDART00000022498
|
tti1
|
TELO2 interacting protein 1 |
| chr4_+_11723852 | 1.17 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr14_-_21661015 | 1.16 |
ENSDART00000189403
ENSDART00000172442 ENSDART00000181913 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr2_+_55365727 | 1.15 |
ENSDART00000162943
|
FP245456.1
|
|
| chr21_+_17051478 | 1.13 |
ENSDART00000047201
ENSDART00000161650 ENSDART00000167298 |
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr4_-_20135406 | 1.12 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr11_-_21586157 | 1.12 |
ENSDART00000190095
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr7_-_30127082 | 1.12 |
ENSDART00000173749
|
alpk3b
|
alpha-kinase 3b |
| chr5_+_36895860 | 1.12 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr7_+_56472585 | 1.11 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr8_+_23147218 | 1.10 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr21_+_3796620 | 1.09 |
ENSDART00000099535
ENSDART00000144515 |
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr11_-_29563437 | 1.09 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr13_-_32906265 | 1.08 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr13_+_28690355 | 1.07 |
ENSDART00000137475
ENSDART00000128246 |
polr1c
|
polymerase (RNA) I polypeptide C |
| chr19_+_33476557 | 1.07 |
ENSDART00000181800
|
triqk
|
triple QxxK/R motif containing |
| chr20_+_53368611 | 1.07 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr25_+_28555584 | 1.07 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr2_+_21982911 | 1.06 |
ENSDART00000190722
ENSDART00000044371 ENSDART00000134912 |
tox
|
thymocyte selection-associated high mobility group box |
| chr3_-_15475067 | 1.05 |
ENSDART00000025324
ENSDART00000139575 |
spns1
|
spinster homolog 1 (Drosophila) |
| chr8_+_23034718 | 1.04 |
ENSDART00000184512
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr23_+_25354856 | 1.04 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr3_+_33440615 | 1.03 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr24_+_14240196 | 1.03 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr22_+_39007533 | 1.03 |
ENSDART00000185958
ENSDART00000129848 |
fam208aa
|
family with sequence similarity 208, member Aa |
| chr25_+_3868438 | 1.03 |
ENSDART00000156438
|
tmem138
|
transmembrane protein 138 |
| chr3_+_53116172 | 1.02 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr17_-_6451801 | 1.01 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr1_+_35494837 | 0.99 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr24_+_27023616 | 0.99 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr23_-_42876596 | 0.99 |
ENSDART00000086156
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr10_+_22918338 | 0.99 |
ENSDART00000167874
ENSDART00000171298 |
zgc:103508
|
zgc:103508 |
| chr12_-_10705916 | 0.98 |
ENSDART00000164038
|
FO704786.1
|
|
| chr14_-_36799280 | 0.97 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr25_-_27843066 | 0.96 |
ENSDART00000179684
ENSDART00000186000 ENSDART00000190065 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr9_+_52411530 | 0.95 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr19_+_619200 | 0.95 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr15_-_4011013 | 0.94 |
ENSDART00000158029
|
CR626884.3
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj1a+foxj1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 1.1 | 2.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.9 | 6.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.8 | 3.4 | GO:0071675 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.8 | 2.4 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.8 | 2.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.7 | 2.2 | GO:0061317 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.7 | 2.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.6 | 2.4 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.6 | 2.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.5 | 2.9 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.5 | 1.4 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.4 | 1.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.4 | 2.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.4 | 1.3 | GO:0045601 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.7 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 2.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 1.6 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.4 | 1.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 1.4 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 1.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 1.7 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.3 | 3.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.3 | 1.5 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.3 | 0.9 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.3 | 3.3 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 1.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 2.0 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.3 | 1.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 2.8 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 3.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.3 | 1.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.3 | 2.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.3 | 3.1 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.3 | 2.1 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.3 | 0.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.3 | 4.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.2 | 1.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.2 | 1.5 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.2 | 4.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 2.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 2.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 1.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 1.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.2 | 0.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 | 0.4 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.2 | 0.6 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 1.7 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 0.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 1.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 2.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 2.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.2 | 2.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.2 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 3.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.7 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 4.3 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 1.3 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.5 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 3.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 2.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.6 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 1.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 2.0 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.9 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 3.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 1.9 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.9 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 2.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 1.8 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 2.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 1.0 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.1 | 1.5 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 3.4 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 2.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.3 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 3.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.8 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 2.0 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.8 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.1 | 1.0 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 1.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 0.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 1.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.2 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 1.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 1.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.7 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 1.2 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:0043091 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 2.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 1.4 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.8 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 1.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.4 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 1.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.7 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.7 | GO:0030518 | intracellular steroid hormone receptor signaling pathway(GO:0030518) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) sulfur compound catabolic process(GO:0044273) |
| 0.0 | 1.6 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 2.0 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 1.2 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.4 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.8 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 2.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 2.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 1.1 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.5 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 3.9 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.7 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 1.2 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 1.1 | GO:0002521 | leukocyte differentiation(GO:0002521) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.8 | 2.4 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.6 | 1.9 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 1.5 | GO:0001534 | radial spoke(GO:0001534) |
| 0.5 | 4.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.4 | 1.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 2.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 2.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.3 | 2.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.3 | 1.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.3 | 2.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.3 | 3.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.3 | 0.8 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 0.9 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 1.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 1.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.2 | 2.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 0.8 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 1.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.2 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 3.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 6.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 4.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 1.5 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.5 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 3.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.6 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.3 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 3.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 4.3 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.2 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 1.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 2.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 4.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 3.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 3.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 7.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 2.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.0 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 4.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.0 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 3.5 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 2.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.9 | 6.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.8 | 2.4 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.8 | 2.3 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 2.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 3.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 2.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.4 | 1.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.4 | 4.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.3 | 2.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.0 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.3 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.3 | 2.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.3 | 2.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 1.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.3 | 1.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 3.9 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.2 | 2.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.0 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 2.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 3.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 1.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 3.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.5 | GO:0047453 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.1 | 0.5 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 2.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.9 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 2.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.7 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.1 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.1 | 2.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 5.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 1.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.8 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.6 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 2.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.9 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 2.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.1 | 5.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 1.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.4 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) |
| 0.1 | 0.3 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.2 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 4.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 3.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 3.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 2.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 3.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 1.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.1 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 3.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 4.5 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 5.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 3.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 8.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 2.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 7.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 2.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.0 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 2.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 2.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 3.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.4 | 2.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.2 | 2.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 3.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 2.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 3.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.0 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 6.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 2.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.4 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.1 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.8 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.9 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.5 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |