Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
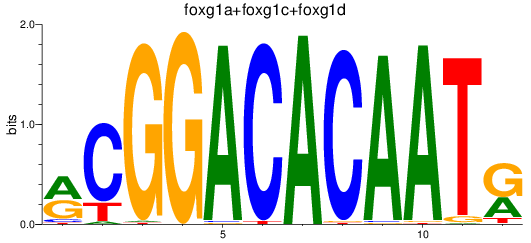
Results for foxg1a+foxg1c+foxg1d
Z-value: 1.28
Transcription factors associated with foxg1a+foxg1c+foxg1d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxg1c
|
ENSDARG00000068380 | forkhead box G1c |
|
foxg1d
|
ENSDARG00000070053 | forkhead box G1d |
|
foxg1a
|
ENSDARG00000070769 | forkhead box G1a |
|
foxg1c
|
ENSDARG00000114414 | forkhead box G1c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxg1a | dr11_v1_chr17_-_29119362_29119362 | -0.83 | 1.2e-05 | Click! |
| foxg1d | dr11_v1_chr13_+_255067_255067 | 0.46 | 4.9e-02 | Click! |
| foxg1c | dr11_v1_chr18_-_40901707_40901707 | 0.27 | 2.6e-01 | Click! |
Activity profile of foxg1a+foxg1c+foxg1d motif
Sorted Z-values of foxg1a+foxg1c+foxg1d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_38088099 | 4.97 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr18_-_38087875 | 4.32 |
ENSDART00000111301
|
luzp2
|
leucine zipper protein 2 |
| chr16_-_13613475 | 4.23 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr19_-_8096984 | 4.21 |
ENSDART00000146987
|
si:dkey-266f7.9
|
si:dkey-266f7.9 |
| chr8_-_51599036 | 3.87 |
ENSDART00000175779
ENSDART00000134614 ENSDART00000098263 |
kctd9a
|
potassium channel tetramerization domain containing 9a |
| chr15_-_30816370 | 3.85 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr25_-_26673570 | 3.80 |
ENSDART00000154917
|
ciartb
|
circadian associated repressor of transcription b |
| chr14_+_2095394 | 3.74 |
ENSDART00000186847
|
CT573264.2
|
|
| chr3_+_23248542 | 3.72 |
ENSDART00000185765
ENSDART00000192332 |
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr14_-_2361692 | 3.55 |
ENSDART00000167696
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.4 |
| chr8_+_19621511 | 3.48 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr18_+_18104235 | 3.20 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr15_-_19250543 | 3.03 |
ENSDART00000092705
ENSDART00000138895 |
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr3_-_15264698 | 3.01 |
ENSDART00000111948
ENSDART00000142594 |
sez6l2
|
seizure related 6 homolog (mouse)-like 2 |
| chr8_+_19621731 | 2.96 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr12_-_17897134 | 2.92 |
ENSDART00000066407
|
nptx2b
|
neuronal pentraxin IIb |
| chr22_-_38543630 | 2.88 |
ENSDART00000172029
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr4_+_1283068 | 2.84 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr23_+_5470967 | 2.70 |
ENSDART00000110522
ENSDART00000149050 |
tulp1a
|
tubby like protein 1a |
| chr11_+_13058613 | 2.61 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr12_+_39685485 | 2.51 |
ENSDART00000163403
|
LO017650.1
|
|
| chr24_+_29381946 | 2.35 |
ENSDART00000189551
|
ntng1a
|
netrin g1a |
| chr17_-_15657029 | 2.27 |
ENSDART00000153925
|
fut9a
|
fucosyltransferase 9a |
| chr25_-_16742438 | 2.18 |
ENSDART00000156091
|
galnt8a.1
|
polypeptide N-acetylgalactosaminyltransferase 8a, tandem duplicate 1 |
| chr4_+_3980247 | 1.93 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr5_+_9360394 | 1.88 |
ENSDART00000124642
|
FP236810.2
|
|
| chr24_+_29382109 | 1.87 |
ENSDART00000184620
ENSDART00000188414 ENSDART00000186132 ENSDART00000191489 |
ntng1a
|
netrin g1a |
| chr4_+_3287819 | 1.85 |
ENSDART00000168633
|
CABZ01085700.1
|
|
| chr15_-_30815826 | 1.76 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr14_-_30452218 | 1.76 |
ENSDART00000011480
|
zdhhc2
|
zinc finger, DHHC-type containing 2 |
| chr7_+_47243564 | 1.62 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr19_-_22541284 | 1.52 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr18_-_5509616 | 1.51 |
ENSDART00000142945
|
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr17_-_24916889 | 1.48 |
ENSDART00000156157
|
si:ch211-195o20.7
|
si:ch211-195o20.7 |
| chr15_-_8763106 | 1.47 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr2_+_30721070 | 1.45 |
ENSDART00000099052
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr7_+_20656942 | 1.37 |
ENSDART00000100898
|
tnfsf12
|
TNF superfamily member 12 |
| chr21_+_43669943 | 1.36 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr7_-_51953807 | 1.30 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr5_+_22791686 | 1.23 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr2_+_30721466 | 1.22 |
ENSDART00000128982
|
si:dkey-94e7.2
|
si:dkey-94e7.2 |
| chr23_+_5977965 | 1.22 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr20_-_14665002 | 1.21 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr20_+_38032143 | 1.11 |
ENSDART00000032161
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr4_-_8902406 | 1.11 |
ENSDART00000192962
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr16_+_41570653 | 1.07 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr1_-_46875493 | 1.05 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr11_-_42554290 | 1.03 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr25_-_35664817 | 0.95 |
ENSDART00000148718
|
lrrk2
|
leucine-rich repeat kinase 2 |
| chr5_-_21044693 | 0.93 |
ENSDART00000140298
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr7_-_51953613 | 0.88 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr14_+_146857 | 0.83 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr1_+_59090583 | 0.80 |
ENSDART00000150658
|
mfap4
|
microfibril associated protein 4 |
| chr19_-_19025998 | 0.71 |
ENSDART00000186156
ENSDART00000163359 ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr24_+_11106402 | 0.67 |
ENSDART00000146697
|
prlh2
|
prolactin releasing hormone 2 |
| chr12_-_43437927 | 0.67 |
ENSDART00000169949
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr19_-_82504 | 0.63 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr1_+_59090743 | 0.63 |
ENSDART00000100199
|
mfap4
|
microfibril associated protein 4 |
| chr24_-_6038025 | 0.56 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr3_+_41726360 | 0.54 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr9_+_2002701 | 0.52 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr7_+_9189547 | 0.51 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr16_-_18702249 | 0.48 |
ENSDART00000191595
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr14_+_12316581 | 0.48 |
ENSDART00000170115
ENSDART00000149757 |
si:ch211-125c23.3
cx31.7
|
si:ch211-125c23.3 connexin 31.7 |
| chr8_-_27656765 | 0.47 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr25_+_21267579 | 0.44 |
ENSDART00000152015
|
fgl2b
|
fibrinogen-like 2b |
| chr7_+_35268880 | 0.42 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr18_+_5510251 | 0.40 |
ENSDART00000007797
|
slc30a4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr5_+_60934534 | 0.40 |
ENSDART00000065025
|
rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr17_-_4252221 | 0.39 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr11_+_583725 | 0.39 |
ENSDART00000189415
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr6_+_11829867 | 0.39 |
ENSDART00000151044
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr19_-_30904590 | 0.37 |
ENSDART00000137633
|
si:ch211-194e15.5
|
si:ch211-194e15.5 |
| chr20_-_54564018 | 0.36 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr5_-_6508250 | 0.29 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr1_-_50710468 | 0.22 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr3_+_23248704 | 0.18 |
ENSDART00000156032
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr11_-_42739148 | 0.06 |
ENSDART00000183584
|
CU459094.2
|
|
| chr8_+_25616946 | 0.04 |
ENSDART00000133983
|
slc38a5a
|
solute carrier family 38, member 5a |
| chr2_-_30721502 | 0.02 |
ENSDART00000132389
|
si:dkey-94e7.1
|
si:dkey-94e7.1 |
| chr6_+_36942966 | 0.01 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxg1a+foxg1c+foxg1d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.4 | 6.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.4 | 5.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 0.9 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 1.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 4.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.8 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.1 | 2.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.7 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 2.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 5.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.5 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 1.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.7 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 2.2 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 3.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.5 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 3.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.9 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 1.0 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 2.3 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 2.6 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.0 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 1.2 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 4.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 5.6 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 1.0 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 6.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 2.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 3.5 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.6 | 1.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.3 | 2.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 2.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 4.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 3.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.4 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.1 | 0.4 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 1.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 2.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 2.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.9 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 4.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 5.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |