Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
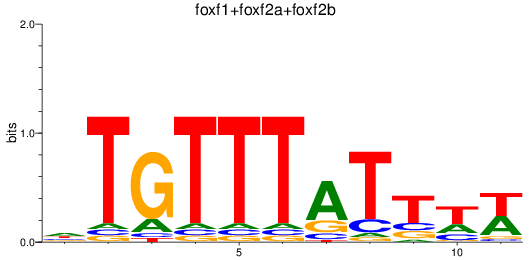
Results for foxf1+foxf2a+foxf2b
Z-value: 0.38
Transcription factors associated with foxf1+foxf2a+foxf2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxf1
|
ENSDARG00000015399 | forkhead box F1 |
|
foxf2a
|
ENSDARG00000017195 | forkhead box F2a |
|
foxf2b
|
ENSDARG00000070389 | forkhead box F2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxf2a | dr11_v1_chr2_-_716426_716426 | -0.63 | 3.6e-03 | Click! |
| foxf1 | dr11_v1_chr18_+_30847237_30847237 | -0.63 | 4.0e-03 | Click! |
| foxf2b | dr11_v1_chr20_+_26690036_26690036 | -0.57 | 1.0e-02 | Click! |
Activity profile of foxf1+foxf2a+foxf2b motif
Sorted Z-values of foxf1+foxf2a+foxf2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_15944245 | 0.75 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr12_+_3912544 | 0.42 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr1_+_51191049 | 0.41 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr1_-_9134045 | 0.37 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr23_-_27152866 | 0.31 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr6_+_34028532 | 0.30 |
ENSDART00000155827
|
si:ch73-185c24.2
|
si:ch73-185c24.2 |
| chr14_-_9713549 | 0.28 |
ENSDART00000193356
ENSDART00000166739 |
si:zfos-2326c3.2
|
si:zfos-2326c3.2 |
| chr9_+_41459759 | 0.26 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr13_-_18011168 | 0.26 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr8_-_13985032 | 0.26 |
ENSDART00000140576
|
igf3
|
insulin-like growth factor 3 |
| chr2_-_38035235 | 0.25 |
ENSDART00000075904
|
cbln5
|
cerebellin 5 |
| chr3_-_49382896 | 0.25 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr7_+_30254652 | 0.25 |
ENSDART00000173711
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr21_-_280769 | 0.25 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr2_+_39618951 | 0.25 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr7_+_15736230 | 0.24 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr22_+_2769236 | 0.24 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr24_+_18299506 | 0.24 |
ENSDART00000172056
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr2_-_42958113 | 0.23 |
ENSDART00000139945
|
oc90
|
otoconin 90 |
| chr2_+_21048661 | 0.23 |
ENSDART00000156876
|
rreb1b
|
ras responsive element binding protein 1b |
| chr20_-_1239596 | 0.22 |
ENSDART00000049063
ENSDART00000140650 |
ankrd6b
|
ankyrin repeat domain 6b |
| chr10_-_11840353 | 0.22 |
ENSDART00000127581
|
trim23
|
tripartite motif containing 23 |
| chr10_+_28428222 | 0.22 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr2_+_44512324 | 0.22 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr11_-_6974022 | 0.22 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr17_-_4245902 | 0.21 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr23_-_32304650 | 0.21 |
ENSDART00000143772
ENSDART00000085642 ENSDART00000188989 |
dgkaa
|
diacylglycerol kinase, alpha a |
| chr23_-_43486714 | 0.21 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr24_+_18299175 | 0.21 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr16_-_27566552 | 0.21 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr9_+_24088062 | 0.20 |
ENSDART00000126198
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr7_+_1579510 | 0.20 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr19_+_43256986 | 0.20 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr16_-_30564319 | 0.20 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr15_-_18432673 | 0.20 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr10_-_36633882 | 0.20 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr11_+_45110865 | 0.19 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr7_-_41554047 | 0.19 |
ENSDART00000174144
|
plxdc2
|
plexin domain containing 2 |
| chr22_+_2144278 | 0.19 |
ENSDART00000162173
ENSDART00000159914 ENSDART00000160192 |
znf1164
|
zinc finger protein 1164 |
| chr8_+_10862353 | 0.19 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr3_+_7771420 | 0.19 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr15_+_3674765 | 0.19 |
ENSDART00000081802
|
NEK3
|
NIMA related kinase 3 |
| chr8_+_29636431 | 0.19 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr1_+_51066671 | 0.19 |
ENSDART00000064007
|
srd5a2a
|
steroid-5-alpha-reductase, alpha polypeptide 2a |
| chr18_+_13077800 | 0.19 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr9_-_7655243 | 0.18 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr4_+_5868034 | 0.18 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr14_-_552036 | 0.18 |
ENSDART00000171317
|
SPATA5
|
spermatogenesis associated 5 |
| chr17_-_5352924 | 0.17 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr2_-_23677422 | 0.17 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr7_+_20629411 | 0.17 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr10_+_573667 | 0.17 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr3_+_49521106 | 0.17 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr19_-_17774875 | 0.17 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr10_+_213878 | 0.17 |
ENSDART00000135903
ENSDART00000138812 |
mpzl1l
|
myelin protein zero-like 1 like |
| chr21_-_3770636 | 0.17 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr20_+_6142433 | 0.17 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr17_-_32743044 | 0.17 |
ENSDART00000135517
|
cenpf
|
centromere protein F |
| chr12_-_26430507 | 0.16 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr15_+_31911989 | 0.16 |
ENSDART00000111472
|
brca2
|
breast cancer 2, early onset |
| chr8_-_6825588 | 0.16 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr13_-_31938512 | 0.16 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr3_-_49138004 | 0.16 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr19_+_43669122 | 0.16 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr19_+_1999838 | 0.16 |
ENSDART00000166669
|
btd
|
biotinidase |
| chr20_-_31496679 | 0.16 |
ENSDART00000153437
ENSDART00000153193 |
sash1a
|
SAM and SH3 domain containing 1a |
| chr22_+_37534094 | 0.16 |
ENSDART00000065239
|
dnajc19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr15_-_7337148 | 0.15 |
ENSDART00000182568
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr1_-_18848955 | 0.15 |
ENSDART00000109294
ENSDART00000146410 |
zgc:195282
|
zgc:195282 |
| chr17_+_7534180 | 0.15 |
ENSDART00000187512
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr4_+_1620102 | 0.15 |
ENSDART00000067444
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr5_-_16351306 | 0.15 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr18_+_40354998 | 0.15 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr5_-_7199998 | 0.15 |
ENSDART00000167316
|
CABZ01087502.1
|
|
| chr6_-_47838481 | 0.15 |
ENSDART00000185284
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr25_+_2776865 | 0.15 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr23_-_25686894 | 0.15 |
ENSDART00000181420
ENSDART00000088208 |
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr3_+_25216790 | 0.15 |
ENSDART00000156544
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr1_+_496268 | 0.15 |
ENSDART00000109415
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr17_+_32622933 | 0.14 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr20_-_48877458 | 0.14 |
ENSDART00000163271
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr7_+_16912837 | 0.14 |
ENSDART00000186956
|
nav2a
|
neuron navigator 2a |
| chr19_+_25971000 | 0.14 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr25_-_13676682 | 0.14 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr13_-_36761379 | 0.14 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_-_53885514 | 0.14 |
ENSDART00000173812
ENSDART00000127144 |
cacna2d2a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2a |
| chr9_+_22780901 | 0.14 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr7_-_59210882 | 0.14 |
ENSDART00000170330
ENSDART00000158996 |
nagk
|
N-acetylglucosamine kinase |
| chr16_-_33104944 | 0.14 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr16_-_35532937 | 0.14 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr10_-_41940302 | 0.14 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr8_-_39859688 | 0.14 |
ENSDART00000019907
|
unc119.1
|
unc-119 homolog 1 |
| chr15_+_16908085 | 0.13 |
ENSDART00000186870
|
ypel2b
|
yippee-like 2b |
| chr9_-_4856767 | 0.13 |
ENSDART00000016814
ENSDART00000138015 |
fmnl2a
|
formin-like 2a |
| chr9_+_21281652 | 0.13 |
ENSDART00000113352
|
lats2
|
large tumor suppressor kinase 2 |
| chr13_-_10620652 | 0.13 |
ENSDART00000135000
ENSDART00000191587 |
si:ch73-54n14.2
camkmt
|
si:ch73-54n14.2 calmodulin-lysine N-methyltransferase |
| chr13_+_25199849 | 0.13 |
ENSDART00000139209
ENSDART00000130876 |
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr6_-_14038804 | 0.13 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr12_+_17436904 | 0.13 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr1_+_54683655 | 0.13 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr13_+_37653851 | 0.13 |
ENSDART00000141988
ENSDART00000126902 ENSDART00000100352 |
phf3
|
PHD finger protein 3 |
| chr8_-_43923788 | 0.13 |
ENSDART00000146152
|
adgrd1
|
adhesion G protein-coupled receptor D1 |
| chr6_-_30683637 | 0.13 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr1_-_58009216 | 0.13 |
ENSDART00000143829
|
nxnl1
|
nucleoredoxin like 1 |
| chr2_+_10134345 | 0.13 |
ENSDART00000100725
|
ahsg2
|
alpha-2-HS-glycoprotein 2 |
| chr17_-_32743249 | 0.13 |
ENSDART00000176858
ENSDART00000189622 |
cenpf
|
centromere protein F |
| chr14_-_2348917 | 0.13 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr24_+_19542323 | 0.13 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr13_-_17860307 | 0.13 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr12_+_27096835 | 0.12 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr6_+_2174082 | 0.12 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr22_+_3184500 | 0.12 |
ENSDART00000176409
ENSDART00000160604 |
ftsj3
|
FtsJ RNA methyltransferase homolog 3 |
| chr11_+_25583950 | 0.12 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr6_+_45918981 | 0.12 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr17_-_27223965 | 0.12 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr23_+_27778670 | 0.12 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr18_-_23874929 | 0.12 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_5356686 | 0.12 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr7_-_24005268 | 0.12 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr12_+_19384615 | 0.12 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr7_-_53117131 | 0.12 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr25_+_6451038 | 0.12 |
ENSDART00000009971
|
snx33
|
sorting nexin 33 |
| chr22_+_31821815 | 0.12 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr22_-_35063526 | 0.12 |
ENSDART00000162211
|
zgc:63733
|
zgc:63733 |
| chr7_-_28611145 | 0.12 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr22_+_30195257 | 0.12 |
ENSDART00000027803
ENSDART00000172496 |
add3a
|
adducin 3 (gamma) a |
| chr23_-_37514493 | 0.12 |
ENSDART00000083267
|
dnajc16
|
DnaJ (Hsp40) homolog, subfamily C, member 16 |
| chr22_-_3152357 | 0.12 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr4_-_17725008 | 0.12 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr20_+_42565049 | 0.12 |
ENSDART00000061101
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr23_+_12916062 | 0.11 |
ENSDART00000144268
|
si:dkey-150i13.2
|
si:dkey-150i13.2 |
| chr23_+_22873415 | 0.11 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr14_-_14607855 | 0.11 |
ENSDART00000162322
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr10_-_7555326 | 0.11 |
ENSDART00000162191
ENSDART00000186945 |
wrn
|
Werner syndrome |
| chr3_-_59297532 | 0.11 |
ENSDART00000187991
|
CABZ01053748.1
|
|
| chr23_+_17417539 | 0.11 |
ENSDART00000182605
|
BX649300.2
|
|
| chr14_+_30910114 | 0.11 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr8_-_31701157 | 0.11 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr4_+_1619584 | 0.11 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr25_+_28555584 | 0.11 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr3_-_5228841 | 0.11 |
ENSDART00000092373
ENSDART00000182438 |
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr9_+_30626416 | 0.11 |
ENSDART00000147813
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr1_-_30510839 | 0.11 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr5_+_66250856 | 0.11 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr6_+_4255319 | 0.11 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr21_-_31143903 | 0.11 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr2_+_36701322 | 0.11 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr22_+_11756040 | 0.11 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr5_+_42259002 | 0.11 |
ENSDART00000083778
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr1_+_23162124 | 0.11 |
ENSDART00000188428
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr23_-_18707418 | 0.11 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr17_+_10593398 | 0.11 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr16_+_52105227 | 0.11 |
ENSDART00000150025
ENSDART00000097863 |
MAN1C1
|
si:ch73-373m9.1 |
| chr1_-_28950366 | 0.11 |
ENSDART00000110270
|
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr5_-_19014589 | 0.11 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr6_-_40922971 | 0.11 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr22_+_38778649 | 0.11 |
ENSDART00000075873
|
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr9_+_45605410 | 0.11 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr7_+_27251376 | 0.11 |
ENSDART00000173521
ENSDART00000173962 |
sox6
|
SRY (sex determining region Y)-box 6 |
| chr19_+_42047427 | 0.11 |
ENSDART00000180225
ENSDART00000145356 |
si:ch211-13c6.2
|
si:ch211-13c6.2 |
| chr25_-_3647277 | 0.10 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr22_-_22301672 | 0.10 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr15_-_20190052 | 0.10 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr15_+_6661343 | 0.10 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr6_+_40922572 | 0.10 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr12_-_5998898 | 0.10 |
ENSDART00000142659
ENSDART00000004896 |
kat7b
|
K(lysine) acetyltransferase 7b |
| chr15_-_7337537 | 0.10 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr20_-_42378865 | 0.10 |
ENSDART00000139912
ENSDART00000015801 |
dcbld1
|
discoidin, CUB and LCCL domain containing 1 |
| chr7_+_1579236 | 0.10 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr21_-_23305561 | 0.10 |
ENSDART00000181815
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr14_+_33723309 | 0.10 |
ENSDART00000132488
|
apln
|
apelin |
| chr8_+_39795918 | 0.10 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr8_+_30456161 | 0.10 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr5_+_69878693 | 0.10 |
ENSDART00000073671
|
ythdc1
|
YTH domain containing 1 |
| chr10_-_3332362 | 0.10 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr21_-_9384374 | 0.10 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr25_+_10830269 | 0.10 |
ENSDART00000175736
|
si:ch211-147g22.5
|
si:ch211-147g22.5 |
| chr17_-_27382826 | 0.10 |
ENSDART00000186657
ENSDART00000155986 ENSDART00000191060 ENSDART00000077608 |
si:ch1073-358c10.1
|
si:ch1073-358c10.1 |
| chr8_-_20862443 | 0.10 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr9_+_55455801 | 0.10 |
ENSDART00000144757
ENSDART00000186543 |
mxra5b
|
matrix-remodelling associated 5b |
| chr10_+_19017146 | 0.10 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr4_+_59711338 | 0.10 |
ENSDART00000150849
|
si:dkey-149m13.4
|
si:dkey-149m13.4 |
| chr17_+_38447473 | 0.10 |
ENSDART00000149007
|
cdan1
|
codanin 1 |
| chr1_+_44710955 | 0.10 |
ENSDART00000131296
ENSDART00000142187 |
ssrp1b
|
structure specific recognition protein 1b |
| chr6_+_32834760 | 0.10 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr7_+_7511914 | 0.10 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr19_+_4968947 | 0.10 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr14_-_26177156 | 0.10 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr24_+_14214831 | 0.10 |
ENSDART00000004664
|
tram1
|
translocation associated membrane protein 1 |
| chr17_-_40956035 | 0.10 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr10_+_8767541 | 0.10 |
ENSDART00000170272
|
itga1
|
integrin, alpha 1 |
| chr11_-_20096018 | 0.10 |
ENSDART00000030420
|
ogfrl2
|
opioid growth factor receptor-like 2 |
| chr12_-_34887943 | 0.10 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr12_+_34854562 | 0.10 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr24_+_18286427 | 0.09 |
ENSDART00000055443
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr15_+_44283723 | 0.09 |
ENSDART00000167722
|
cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr18_-_24988645 | 0.09 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr23_-_33558161 | 0.09 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr17_+_16046314 | 0.09 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_+_29726428 | 0.09 |
ENSDART00000143183
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxf1+foxf2a+foxf2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.2 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.1 | 0.4 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.1 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.4 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.2 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.0 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.1 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.1 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.1 | GO:0070883 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |