Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
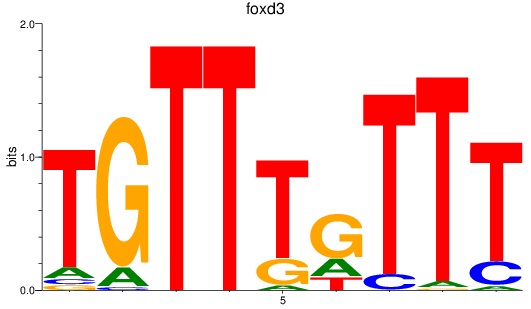
Results for foxd3
Z-value: 1.37
Transcription factors associated with foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd3
|
ENSDARG00000021032 | forkhead box D3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd3 | dr11_v1_chr6_-_32093830_32093830 | 0.74 | 2.6e-04 | Click! |
Activity profile of foxd3 motif
Sorted Z-values of foxd3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_48049202 | 4.24 |
ENSDART00000027158
|
psmd3
|
proteasome 26S subunit, non-ATPase 3 |
| chr7_-_28611145 | 3.12 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr19_+_15443353 | 2.88 |
ENSDART00000135923
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr23_-_27152866 | 2.86 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr16_+_19537073 | 2.67 |
ENSDART00000190590
|
sp8b
|
sp8 transcription factor b |
| chr2_-_44777592 | 2.66 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr17_-_114121 | 2.48 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr7_+_21787507 | 2.37 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr12_+_28888975 | 2.15 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr6_-_25201810 | 2.13 |
ENSDART00000168683
|
lrrc8c
|
leucine rich repeat containing 8 VRAC subunit C |
| chr2_-_42958113 | 2.07 |
ENSDART00000139945
|
oc90
|
otoconin 90 |
| chr7_+_15736230 | 2.03 |
ENSDART00000109942
|
mctp2b
|
multiple C2 domains, transmembrane 2b |
| chr14_-_16082806 | 2.03 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr4_+_15944245 | 2.01 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr5_+_36513605 | 1.96 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr2_-_42958619 | 1.92 |
ENSDART00000144317
|
oc90
|
otoconin 90 |
| chr7_-_41554047 | 1.88 |
ENSDART00000174144
|
plxdc2
|
plexin domain containing 2 |
| chr12_-_17655683 | 1.87 |
ENSDART00000066411
|
dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr2_+_11685742 | 1.87 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr19_+_15443063 | 1.82 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_+_24994705 | 1.81 |
ENSDART00000129211
|
zgc:92107
|
zgc:92107 |
| chr25_-_14433503 | 1.80 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr23_-_27647769 | 1.74 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr13_+_5978809 | 1.69 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr14_-_26704829 | 1.69 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr4_-_9909371 | 1.68 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr12_+_3912544 | 1.67 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr16_+_42829735 | 1.65 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr15_-_44052927 | 1.63 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr9_-_6661657 | 1.62 |
ENSDART00000133178
ENSDART00000113914 ENSDART00000061593 |
pou3f3a
|
POU class 3 homeobox 3a |
| chr5_-_22027357 | 1.59 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr9_+_15893093 | 1.59 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr6_-_14038804 | 1.57 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr2_-_10098191 | 1.55 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr24_+_36317544 | 1.54 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr3_+_23743139 | 1.49 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr2_-_25143373 | 1.44 |
ENSDART00000160108
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr11_-_6070192 | 1.43 |
ENSDART00000162776
|
babam1
|
BRISC and BRCA1 A complex member 1 |
| chr19_+_25971000 | 1.41 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr18_-_23875370 | 1.39 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr18_-_16922905 | 1.39 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr17_-_4245902 | 1.39 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr18_-_23875219 | 1.35 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_22067570 | 1.35 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr6_+_39370587 | 1.35 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr5_+_61476014 | 1.33 |
ENSDART00000050906
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr20_-_9980318 | 1.32 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr10_-_10863936 | 1.32 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr1_-_33647138 | 1.32 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr18_+_40354998 | 1.31 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr9_+_38458193 | 1.30 |
ENSDART00000008053
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr8_+_47850073 | 1.30 |
ENSDART00000083395
|
plod1a
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1a |
| chr6_-_40922971 | 1.30 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr8_-_20138054 | 1.28 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr13_-_11967769 | 1.28 |
ENSDART00000158369
|
ARL3 (1 of many)
|
zgc:110197 |
| chr25_+_16646113 | 1.27 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr25_-_18330503 | 1.23 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr17_+_32622933 | 1.23 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr16_+_40131473 | 1.23 |
ENSDART00000155421
ENSDART00000134732 ENSDART00000138699 |
cenpw
si:ch211-195p4.4
|
centromere protein W si:ch211-195p4.4 |
| chr23_-_22113455 | 1.22 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr10_+_22775253 | 1.21 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr7_+_24023653 | 1.21 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr17_-_27419319 | 1.21 |
ENSDART00000127043
|
ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr8_-_37043900 | 1.20 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr14_-_21660548 | 1.19 |
ENSDART00000161713
ENSDART00000089845 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr25_+_16312258 | 1.19 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr10_-_10864331 | 1.19 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr4_+_14981854 | 1.19 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr8_+_26293673 | 1.18 |
ENSDART00000144977
|
mgll
|
monoglyceride lipase |
| chr21_+_45268112 | 1.17 |
ENSDART00000157136
|
tcf7
|
transcription factor 7 |
| chr1_+_9153141 | 1.16 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr21_+_45267589 | 1.16 |
ENSDART00000182963
ENSDART00000155681 ENSDART00000192632 |
tcf7
|
transcription factor 7 |
| chr25_+_18965430 | 1.16 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr12_+_13118540 | 1.16 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr5_-_8171625 | 1.13 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr7_+_16991711 | 1.13 |
ENSDART00000173660
|
nav2a
|
neuron navigator 2a |
| chr7_-_54678289 | 1.13 |
ENSDART00000170774
|
ccnd1
|
cyclin D1 |
| chr20_+_13883131 | 1.12 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr9_+_30633184 | 1.12 |
ENSDART00000191310
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr17_+_16423721 | 1.12 |
ENSDART00000064233
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr1_+_11107688 | 1.12 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr8_-_25566347 | 1.11 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr5_-_22052852 | 1.11 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr10_-_31563049 | 1.11 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr15_-_7337537 | 1.10 |
ENSDART00000161613
|
SLC7A1 (1 of many)
|
high affinity cationic amino acid transporter 1 |
| chr4_+_69150355 | 1.09 |
ENSDART00000172364
ENSDART00000184374 |
si:ch211-209j12.2
|
si:ch211-209j12.2 |
| chr11_-_6974022 | 1.09 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr2_+_23790748 | 1.08 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr4_-_16545085 | 1.07 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr9_+_48761455 | 1.07 |
ENSDART00000139631
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr16_+_19536614 | 1.06 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr23_+_31245395 | 1.06 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr12_+_27096835 | 1.05 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr3_-_60175470 | 1.05 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr9_-_23147026 | 1.05 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr19_-_29303788 | 1.05 |
ENSDART00000112167
|
srfbp1
|
serum response factor binding protein 1 |
| chr11_-_18323059 | 1.05 |
ENSDART00000182590
|
SFMBT1
|
Scm like with four mbt domains 1 |
| chr2_+_58841181 | 1.04 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr8_+_29593986 | 1.04 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr4_-_67589158 | 1.04 |
ENSDART00000184361
|
BX088712.2
|
|
| chr3_+_28502419 | 1.04 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr2_+_33926606 | 1.04 |
ENSDART00000111430
|
kif2c
|
kinesin family member 2C |
| chr8_+_26432677 | 1.03 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr1_-_55166511 | 1.03 |
ENSDART00000150430
ENSDART00000035725 |
pane1
|
proliferation associated nuclear element |
| chr13_+_11828516 | 1.03 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr12_-_28910419 | 1.03 |
ENSDART00000153278
ENSDART00000152937 |
CCDC189
|
si:ch73-81k8.2 |
| chr1_-_33645967 | 1.02 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr15_+_19990068 | 1.02 |
ENSDART00000154033
ENSDART00000054428 |
zgc:112083
|
zgc:112083 |
| chr18_-_19005919 | 1.02 |
ENSDART00000129776
|
ints14
|
integrator complex subunit 14 |
| chr12_-_10476448 | 1.02 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr25_+_2776865 | 1.01 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr9_+_38457806 | 1.01 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr1_-_31534089 | 1.01 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr14_-_16772452 | 1.00 |
ENSDART00000137493
|
mrnip
|
MRN complex interacting protein |
| chr23_-_17003533 | 1.00 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr8_+_39570615 | 1.00 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr2_+_15069011 | 1.00 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr2_+_13710439 | 0.99 |
ENSDART00000155712
|
ebna1bp2
|
EBNA1 binding protein 2 |
| chr1_-_26071535 | 0.99 |
ENSDART00000185187
|
pdcd4a
|
programmed cell death 4a |
| chr13_-_42400647 | 0.99 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr9_-_29844596 | 0.99 |
ENSDART00000138574
|
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr22_-_10591876 | 0.98 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr8_-_16725573 | 0.98 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr9_+_27720428 | 0.97 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr2_+_33926911 | 0.97 |
ENSDART00000109849
ENSDART00000135884 |
kif2c
|
kinesin family member 2C |
| chr7_-_7766920 | 0.97 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr25_-_11016675 | 0.97 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr9_-_30220818 | 0.97 |
ENSDART00000140929
|
si:dkey-100n23.3
|
si:dkey-100n23.3 |
| chr4_+_1620102 | 0.96 |
ENSDART00000067444
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr24_+_41690545 | 0.96 |
ENSDART00000160069
|
lama1
|
laminin, alpha 1 |
| chr24_+_18299506 | 0.95 |
ENSDART00000172056
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr5_+_67390645 | 0.95 |
ENSDART00000014822
|
ebf2
|
early B cell factor 2 |
| chr13_-_35459928 | 0.95 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr10_+_19017146 | 0.95 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr11_+_18612421 | 0.95 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr5_+_25762271 | 0.93 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr23_+_22873415 | 0.93 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr18_-_23874929 | 0.93 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr15_-_14038227 | 0.92 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr11_+_7528599 | 0.92 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr7_+_56577906 | 0.92 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr19_-_47782916 | 0.92 |
ENSDART00000063337
|
cdca8
|
cell division cycle associated 8 |
| chr9_+_41459759 | 0.92 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr18_+_26429428 | 0.92 |
ENSDART00000142686
|
blm
|
Bloom syndrome, RecQ helicase-like |
| chr23_-_3758637 | 0.91 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr2_-_10877228 | 0.91 |
ENSDART00000138718
ENSDART00000034246 |
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr4_-_64141714 | 0.91 |
ENSDART00000128628
|
BX914205.3
|
|
| chr16_-_30564319 | 0.91 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr15_-_20024205 | 0.90 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr20_+_29209615 | 0.90 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr15_-_41689684 | 0.90 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr7_-_51102479 | 0.90 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr17_-_20236228 | 0.90 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr12_+_27537357 | 0.90 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr3_-_34561624 | 0.89 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr9_-_30054178 | 0.89 |
ENSDART00000139863
|
dcbld2
|
discoidin, CUB and LCCL domain containing 2 |
| chr17_+_37932433 | 0.89 |
ENSDART00000185349
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr18_-_45761868 | 0.89 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr22_+_2417105 | 0.89 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr4_-_13921185 | 0.89 |
ENSDART00000143202
ENSDART00000080334 |
yaf2
|
YY1 associated factor 2 |
| chr10_+_20070178 | 0.89 |
ENSDART00000027612
ENSDART00000145264 ENSDART00000172713 |
xpo7
|
exportin 7 |
| chr14_+_18782610 | 0.89 |
ENSDART00000164468
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr22_-_4813345 | 0.88 |
ENSDART00000190650
|
CU570881.2
|
|
| chr19_-_2822372 | 0.88 |
ENSDART00000109130
ENSDART00000122385 |
recql4
|
RecQ helicase-like 4 |
| chr17_-_42218652 | 0.88 |
ENSDART00000081396
ENSDART00000190007 |
nkx2.2a
|
NK2 homeobox 2a |
| chr12_-_4301234 | 0.88 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr7_+_52712807 | 0.88 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr17_-_20228610 | 0.88 |
ENSDART00000125758
|
ebf3b
|
early B cell factor 3b |
| chr14_-_26498196 | 0.88 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr18_+_20226843 | 0.88 |
ENSDART00000100632
|
tle3a
|
transducin-like enhancer of split 3a |
| chr17_-_16422654 | 0.88 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr7_-_24995631 | 0.88 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr24_+_19542323 | 0.87 |
ENSDART00000140379
ENSDART00000142830 |
sulf1
|
sulfatase 1 |
| chr7_-_37917517 | 0.87 |
ENSDART00000173795
|
heatr3
|
HEAT repeat containing 3 |
| chr23_-_23160088 | 0.87 |
ENSDART00000141853
|
noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr8_-_7567815 | 0.86 |
ENSDART00000132536
|
hcfc1b
|
host cell factor C1b |
| chr16_-_39131666 | 0.86 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr8_+_8927870 | 0.86 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr9_+_41024973 | 0.86 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr13_+_25200105 | 0.85 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr9_-_30555725 | 0.85 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr6_+_43426599 | 0.85 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr7_-_59159253 | 0.85 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr19_-_10323845 | 0.84 |
ENSDART00000151259
ENSDART00000151821 |
u2af2b
|
U2 small nuclear RNA auxiliary factor 2b |
| chr5_-_14500622 | 0.84 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr19_+_16064439 | 0.84 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr5_+_41996889 | 0.84 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr25_-_13789955 | 0.84 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr23_-_10696626 | 0.83 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr1_+_26676758 | 0.83 |
ENSDART00000152299
|
si:dkey-25o16.4
|
si:dkey-25o16.4 |
| chr2_+_37838259 | 0.83 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr19_-_47782586 | 0.83 |
ENSDART00000177126
|
cdca8
|
cell division cycle associated 8 |
| chr11_-_25829712 | 0.83 |
ENSDART00000103549
|
skib
|
v-ski avian sarcoma viral oncogene homolog b |
| chr8_+_50742975 | 0.83 |
ENSDART00000155664
ENSDART00000160612 |
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr25_-_31423493 | 0.82 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr11_+_31864921 | 0.82 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr22_+_38978084 | 0.82 |
ENSDART00000025482
|
arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr6_+_41446541 | 0.82 |
ENSDART00000029553
ENSDART00000128756 ENSDART00000144864 |
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr3_-_55650417 | 0.82 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr12_-_11349899 | 0.82 |
ENSDART00000079645
|
zgc:174164
|
zgc:174164 |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.7 | 3.7 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.7 | 2.2 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.7 | 2.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 2.3 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.5 | 1.6 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.5 | 1.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.5 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.2 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.4 | 1.2 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.4 | 2.0 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 1.9 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 1.7 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.3 | 1.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.3 | 1.3 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 1.0 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 1.5 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 1.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.3 | 4.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 1.4 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.3 | 0.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 1.1 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.3 | 3.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.3 | 1.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.3 | 1.0 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 1.5 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 2.5 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.2 | 0.7 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.2 | 1.7 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 1.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.2 | 2.4 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.2 | 0.7 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 0.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.2 | 0.8 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.2 | 0.4 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 2.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 3.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.6 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.2 | 1.9 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 0.6 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 2.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.5 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.2 | 0.7 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 1.4 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.2 | 1.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 1.2 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.2 | 0.7 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.4 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.2 | 0.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 4.5 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.2 | 0.5 | GO:0061194 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.2 | 0.3 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 | 1.3 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.2 | 1.0 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 0.5 | GO:0046822 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleocytoplasmic transport(GO:0046822) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 0.5 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 0.3 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.2 | 1.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.2 | 1.9 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 1.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.9 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.9 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.4 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 0.4 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.1 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 0.7 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.8 | GO:0046637 | regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043370) regulation of alpha-beta T cell differentiation(GO:0046637) |
| 0.1 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 1.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 1.6 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 1.4 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.4 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.8 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.1 | 0.9 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.9 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.4 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.4 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.7 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.4 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.5 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.1 | 1.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.7 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.5 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.1 | 0.7 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:0071025 | RNA surveillance(GO:0071025) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.3 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.1 | 0.3 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 1.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.6 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.8 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.9 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.7 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.4 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.5 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.7 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.1 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.1 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 1.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.3 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 1.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.9 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.1 | 0.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.9 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 1.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 2.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.7 | GO:0060173 | limb development(GO:0060173) |
| 0.1 | 1.0 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 1.0 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 1.0 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.4 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 2.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 0.8 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.1 | 1.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 2.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 2.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.3 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 1.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.5 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 1.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.6 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.5 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 0.3 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 0.2 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 1.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.3 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.5 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 0.6 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 0.1 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.1 | 0.4 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.2 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.3 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.1 | GO:0044785 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.9 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.4 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.8 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 0.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.4 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0044319 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) |
| 0.1 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.4 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.1 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 1.1 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.2 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.0 | 0.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 1.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0045638 | negative regulation of myeloid cell differentiation(GO:0045638) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 3.2 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.6 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.1 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.0 | 1.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.2 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 2.2 | GO:0050922 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.0 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.9 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 1.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.6 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.9 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.9 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.0 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.0 | 0.8 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.2 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0048538 | thymus development(GO:0048538) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 2.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 1.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.7 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 2.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 1.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.3 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 1.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.9 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 1.6 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 1.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.5 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 2.1 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 0.4 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 1.5 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.0 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.4 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.2 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.7 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.0 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.8 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 1.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:2001270 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.6 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.7 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.4 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.0 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.4 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 1.1 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.0 | 0.5 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.2 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 1.1 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 2.1 | GO:0061008 | hepaticobiliary system development(GO:0061008) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.7 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.7 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.4 | GO:0035924 | cellular response to vascular endothelial growth factor stimulus(GO:0035924) |
| 0.0 | 0.4 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.8 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.9 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.4 | GO:0046463 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.1 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 1.2 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.0 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) piRNA biosynthetic process(GO:1990511) |
| 0.0 | 1.1 | GO:0006402 | mRNA catabolic process(GO:0006402) |
| 0.0 | 0.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.5 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 1.2 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.0 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.0 | 1.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.9 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.4 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 1.9 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.1 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.2 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 1.0 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.5 | 2.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.4 | 4.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.3 | 1.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.3 | 1.8 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 0.9 | GO:0030689 | Noc complex(GO:0030689) |
| 0.3 | 1.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 1.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.8 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.2 | 0.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 0.5 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 2.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 1.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.2 | 1.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 1.4 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 0.7 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 1.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 1.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.2 | 0.6 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.9 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.5 | GO:0070743 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.8 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.1 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.4 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.6 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.8 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.7 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 0.6 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.9 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.4 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 1.9 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 6.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.7 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 5.0 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.6 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.3 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.2 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.8 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.6 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 1.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 7.2 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.0 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.4 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 2.5 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.0 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 4.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 3.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.3 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 5.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.8 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 7.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 2.0 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 2.6 | GO:0005764 | lysosome(GO:0005764) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.5 | 3.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.4 | 1.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.4 | 2.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.4 | 2.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 0.9 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 1.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.3 | 1.0 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 0.6 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.8 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 1.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.2 | 1.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 0.5 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 1.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 3.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 0.5 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.2 | 0.6 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.6 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 1.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.7 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.2 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.5 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.0 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.4 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 2.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.8 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 1.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.7 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.3 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 3.8 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 0.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.6 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.6 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.7 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.3 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.1 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.3 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.2 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.2 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.0 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.1 | 0.4 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.5 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 3.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 3.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 5.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 1.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.3 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.6 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 8.8 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.4 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 3.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 4.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 41.7 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.4 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.6 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 1.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.5 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.0 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 4.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 1.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.2 | 7.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 2.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.7 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 5.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.2 | 2.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 2.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 6.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 1.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.5 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.1 | 4.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 7.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.8 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 2.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.1 | 3.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 2.3 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.1 | 1.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 3.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.5 | REACTOME S PHASE | Genes involved in S Phase |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 1.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.6 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |