Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
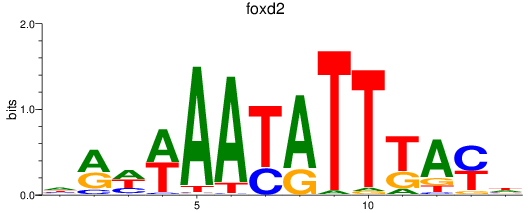
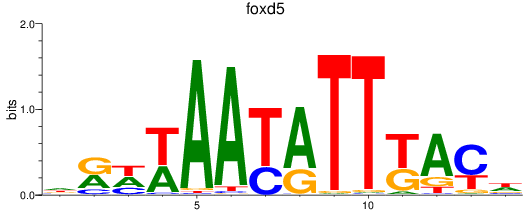
Results for foxd2_foxd5
Z-value: 1.45
Transcription factors associated with foxd2_foxd5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd2
|
ENSDARG00000058133 | forkhead box D2 |
|
foxd5
|
ENSDARG00000042485 | forkhead box D5 |
|
foxd5
|
ENSDARG00000109712 | forkhead box D5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd2 | dr11_v1_chr8_+_19674369_19674369 | -0.92 | 2.5e-08 | Click! |
| foxd5 | dr11_v1_chr8_+_30452945_30452945 | -0.65 | 2.4e-03 | Click! |
Activity profile of foxd2_foxd5 motif
Sorted Z-values of foxd2_foxd5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_3237561 | 6.28 |
ENSDART00000164665
|
si:ch1073-13h15.3
|
si:ch1073-13h15.3 |
| chr12_-_22524388 | 5.09 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr4_+_7841627 | 4.49 |
ENSDART00000037997
|
ucmaa
|
upper zone of growth plate and cartilage matrix associated a |
| chr21_+_40092301 | 4.38 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr15_-_34408777 | 3.97 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr4_+_12013043 | 3.88 |
ENSDART00000130692
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr10_+_36650222 | 3.32 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr15_-_37829160 | 3.26 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr8_+_16004154 | 2.98 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_-_21280373 | 2.89 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr6_+_41186320 | 2.86 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr25_+_24250247 | 2.77 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr7_-_12909352 | 2.75 |
ENSDART00000172901
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr16_-_13388821 | 2.75 |
ENSDART00000144062
|
grin2db
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, b |
| chr25_+_29160102 | 2.49 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr20_+_34717403 | 2.45 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr9_+_34127005 | 2.41 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr25_+_4760489 | 2.41 |
ENSDART00000167399
|
FP245455.1
|
|
| chr5_+_26795465 | 2.39 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr23_-_44219902 | 2.33 |
ENSDART00000185874
|
zgc:158659
|
zgc:158659 |
| chr6_-_24053404 | 2.25 |
ENSDART00000168511
|
si:dkey-44g17.6
|
si:dkey-44g17.6 |
| chr14_-_4121052 | 2.25 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr25_+_19149241 | 2.23 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr16_-_44349845 | 2.18 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr20_+_36233873 | 2.08 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr2_-_9748039 | 2.04 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr1_-_37087966 | 2.03 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr18_-_6803424 | 2.00 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr1_-_30473422 | 1.96 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr24_-_35707552 | 1.95 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_-_4120636 | 1.91 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr10_+_26972755 | 1.91 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr12_+_40905427 | 1.88 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr11_+_6295370 | 1.80 |
ENSDART00000139882
|
ranbp3a
|
RAN binding protein 3a |
| chr15_+_45563656 | 1.77 |
ENSDART00000157501
|
cldn15lb
|
claudin 15-like b |
| chr14_+_46410766 | 1.77 |
ENSDART00000032342
|
anxa5a
|
annexin A5a |
| chr6_+_40671336 | 1.75 |
ENSDART00000111639
ENSDART00000186617 |
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr3_-_28828242 | 1.74 |
ENSDART00000151445
|
si:ch211-76l23.4
|
si:ch211-76l23.4 |
| chr13_-_30662403 | 1.69 |
ENSDART00000012457
|
si:dkey-275b16.2
|
si:dkey-275b16.2 |
| chr18_+_14277003 | 1.69 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr11_-_37880492 | 1.65 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr10_+_33744098 | 1.65 |
ENSDART00000147775
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr6_-_55864687 | 1.65 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr14_-_33894915 | 1.64 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr7_+_21841037 | 1.47 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr3_+_34683096 | 1.47 |
ENSDART00000084432
|
dusp3b
|
dual specificity phosphatase 3b |
| chr20_+_43083745 | 1.46 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr13_+_32446169 | 1.43 |
ENSDART00000143325
|
nt5c1ba
|
5'-nucleotidase, cytosolic IB a |
| chr5_-_45634675 | 1.40 |
ENSDART00000168534
|
NPFFR2
|
neuropeptide FF receptor 2a |
| chr8_+_23142946 | 1.39 |
ENSDART00000152933
|
si:ch211-196c10.13
|
si:ch211-196c10.13 |
| chr20_-_9963713 | 1.34 |
ENSDART00000104234
|
gjd2b
|
gap junction protein delta 2b |
| chr16_-_29334672 | 1.33 |
ENSDART00000162835
|
bcan
|
brevican |
| chr17_+_30448452 | 1.30 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr14_-_4556896 | 1.30 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr18_-_16181952 | 1.25 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr18_-_17077419 | 1.21 |
ENSDART00000148714
|
il17c
|
interleukin 17c |
| chr15_+_45563491 | 1.21 |
ENSDART00000191169
|
cldn15lb
|
claudin 15-like b |
| chr17_-_29311835 | 1.19 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr10_+_26973063 | 1.18 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr14_+_11457500 | 1.18 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr5_+_32206378 | 1.18 |
ENSDART00000126873
ENSDART00000051361 |
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr2_-_4787566 | 1.16 |
ENSDART00000160663
ENSDART00000157808 |
tnk2b
|
tyrosine kinase, non-receptor, 2b |
| chr14_+_11458044 | 1.14 |
ENSDART00000186425
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr6_-_40697585 | 1.14 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr8_+_16004551 | 1.12 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr19_+_9186175 | 1.08 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr12_-_36260532 | 1.07 |
ENSDART00000022533
|
kcnj2a
|
potassium inwardly-rectifying channel, subfamily J, member 2a |
| chr19_+_32807236 | 1.05 |
ENSDART00000008698
|
stk3
|
serine/threonine kinase 3 (STE20 homolog, yeast) |
| chr7_-_35515931 | 1.04 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr20_-_3166168 | 1.01 |
ENSDART00000134137
|
si:ch73-212j7.3
|
si:ch73-212j7.3 |
| chr14_-_41556720 | 1.00 |
ENSDART00000149244
|
itga6l
|
integrin, alpha 6, like |
| chr22_+_1779401 | 0.98 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr5_+_13385837 | 0.95 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr6_+_2097690 | 0.94 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr2_-_37960688 | 0.93 |
ENSDART00000055565
|
cbln14
|
cerebellin 14 |
| chr17_-_29312506 | 0.93 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr12_+_2648043 | 0.88 |
ENSDART00000082220
|
gdf2
|
growth differentiation factor 2 |
| chr4_+_74551606 | 0.88 |
ENSDART00000174339
|
kcna1b
|
potassium voltage-gated channel, shaker-related subfamily, member 1b |
| chr21_-_18993110 | 0.88 |
ENSDART00000144086
|
si:ch211-222n4.6
|
si:ch211-222n4.6 |
| chr24_+_5840258 | 0.88 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr1_-_10473630 | 0.87 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr16_-_16619854 | 0.86 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr7_+_65387374 | 0.86 |
ENSDART00000188680
|
CLEC3A
|
C-type lectin domain family 3 member A |
| chr13_+_32740509 | 0.85 |
ENSDART00000076423
ENSDART00000160138 |
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr11_-_25730416 | 0.84 |
ENSDART00000191612
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr9_+_22677503 | 0.82 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr24_+_8904741 | 0.80 |
ENSDART00000140924
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr14_+_21107032 | 0.80 |
ENSDART00000138319
ENSDART00000139103 ENSDART00000184735 |
aldob
|
aldolase b, fructose-bisphosphate |
| chr6_+_9870192 | 0.80 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr13_+_16521898 | 0.80 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr9_+_52398531 | 0.80 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr18_+_13837746 | 0.79 |
ENSDART00000169552
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr19_-_31522625 | 0.78 |
ENSDART00000158438
ENSDART00000035049 |
necab1
|
N-terminal EF-hand calcium binding protein 1 |
| chr4_+_12013642 | 0.78 |
ENSDART00000067281
|
cry1aa
|
cryptochrome circadian clock 1aa |
| chr19_+_32201991 | 0.73 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr16_+_11724230 | 0.72 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr23_-_42810664 | 0.70 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr8_+_26410197 | 0.69 |
ENSDART00000145836
ENSDART00000053447 |
ifrd2
|
interferon-related developmental regulator 2 |
| chr20_+_27647436 | 0.69 |
ENSDART00000180557
ENSDART00000185628 |
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr5_-_67750907 | 0.68 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr11_+_41858807 | 0.67 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr20_+_36234335 | 0.67 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr10_-_25543227 | 0.66 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr23_-_35790235 | 0.66 |
ENSDART00000142369
ENSDART00000141141 ENSDART00000011004 |
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr20_-_30369598 | 0.66 |
ENSDART00000144549
|
allc
|
allantoicase |
| chr18_+_19419120 | 0.65 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr15_+_24691088 | 0.65 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr21_+_27416284 | 0.64 |
ENSDART00000077593
ENSDART00000108763 |
cfb
|
complement factor B |
| chr17_-_19963718 | 0.63 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr9_-_16109001 | 0.63 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr22_-_11493236 | 0.61 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr4_+_57093908 | 0.61 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr8_+_9149436 | 0.60 |
ENSDART00000064090
|
pnck
|
pregnancy up-regulated nonubiquitous CaM kinase |
| chr12_-_20120702 | 0.60 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr15_+_24212847 | 0.59 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr8_+_7975745 | 0.59 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr24_-_22756508 | 0.59 |
ENSDART00000035409
ENSDART00000146247 |
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr10_-_35236949 | 0.59 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr5_+_4575800 | 0.58 |
ENSDART00000180952
|
cst14b.2
|
cystatin 14b, tandem duplicate 2 |
| chr20_-_25436389 | 0.58 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr9_+_45789887 | 0.57 |
ENSDART00000135202
|
si:dkey-34f9.3
|
si:dkey-34f9.3 |
| chr22_-_9728208 | 0.56 |
ENSDART00000185962
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr7_+_53156810 | 0.56 |
ENSDART00000189816
|
cdh29
|
cadherin 29 |
| chr3_-_45848043 | 0.54 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr7_+_35245607 | 0.54 |
ENSDART00000193422
ENSDART00000173888 |
amfrb
|
autocrine motility factor receptor b |
| chr19_+_7043634 | 0.52 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr1_+_44582369 | 0.52 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
| chr3_-_34724879 | 0.52 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr3_+_25216790 | 0.51 |
ENSDART00000156544
|
c1qtnf6a
|
C1q and TNF related 6a |
| chr5_-_69707787 | 0.51 |
ENSDART00000108820
ENSDART00000149692 |
dguok
|
deoxyguanosine kinase |
| chr3_-_45848257 | 0.50 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr14_-_33613794 | 0.50 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr6_-_40098641 | 0.50 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr11_-_13152215 | 0.49 |
ENSDART00000160989
ENSDART00000158239 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr17_+_11507131 | 0.49 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr8_-_47329755 | 0.48 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr12_+_28986308 | 0.48 |
ENSDART00000153178
|
si:rp71-1c10.8
|
si:rp71-1c10.8 |
| chr15_+_23911655 | 0.46 |
ENSDART00000156304
|
si:ch1073-145m9.1
|
si:ch1073-145m9.1 |
| chr15_-_16121496 | 0.44 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr12_+_29173523 | 0.43 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr18_-_5111449 | 0.42 |
ENSDART00000046902
|
pdcd10a
|
programmed cell death 10a |
| chr2_-_985417 | 0.42 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr5_+_22970617 | 0.42 |
ENSDART00000192859
|
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr20_-_49889111 | 0.42 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr12_-_5455936 | 0.41 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr17_+_12942634 | 0.40 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr17_-_24564674 | 0.40 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr19_+_32856907 | 0.37 |
ENSDART00000148232
|
rpl30
|
ribosomal protein L30 |
| chr24_+_7631797 | 0.36 |
ENSDART00000187464
|
cavin1b
|
caveolae associated protein 1b |
| chr2_-_48171112 | 0.36 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr16_+_23975930 | 0.35 |
ENSDART00000147858
ENSDART00000144347 ENSDART00000115270 |
apoc4
|
apolipoprotein C-IV |
| chr4_-_7212875 | 0.35 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr1_+_532766 | 0.35 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr18_-_17077596 | 0.35 |
ENSDART00000111821
|
il17c
|
interleukin 17c |
| chr17_+_12942021 | 0.35 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr2_+_23007675 | 0.34 |
ENSDART00000163649
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr10_+_39248911 | 0.34 |
ENSDART00000170079
ENSDART00000167974 |
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr6_+_40437987 | 0.34 |
ENSDART00000136487
|
ghrl
|
ghrelin/obestatin prepropeptide |
| chr3_+_36424055 | 0.33 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr20_-_51267185 | 0.33 |
ENSDART00000158350
ENSDART00000023064 |
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr22_+_1586060 | 0.32 |
ENSDART00000160793
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr18_-_37007294 | 0.31 |
ENSDART00000088309
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr16_-_52966811 | 0.31 |
ENSDART00000153758
ENSDART00000009256 |
brd9
|
bromodomain containing 9 |
| chr15_+_32867420 | 0.30 |
ENSDART00000159442
|
dclk1b
|
doublecortin-like kinase 1b |
| chr2_-_127945 | 0.30 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr7_+_13039426 | 0.29 |
ENSDART00000171777
|
syt7b
|
synaptotagmin VIIb |
| chr1_-_656693 | 0.28 |
ENSDART00000170483
ENSDART00000166786 |
appa
|
amyloid beta (A4) precursor protein a |
| chr15_-_5157572 | 0.27 |
ENSDART00000174192
|
or128-10
|
odorant receptor, family E, subfamily 128, member 10 |
| chr22_-_15562933 | 0.27 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr18_-_37007061 | 0.27 |
ENSDART00000136432
|
map3k10
|
mitogen-activated protein kinase kinase kinase 10 |
| chr16_+_46492994 | 0.26 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr21_-_39327223 | 0.26 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr11_-_40742424 | 0.25 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr8_+_19624589 | 0.25 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr19_-_42238440 | 0.25 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr11_-_13152524 | 0.25 |
ENSDART00000181440
ENSDART00000191997 |
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr10_+_34315719 | 0.24 |
ENSDART00000135303
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr6_+_24661736 | 0.24 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr17_+_8292892 | 0.23 |
ENSDART00000125728
|
si:ch211-236p5.3
|
si:ch211-236p5.3 |
| chr15_+_17623115 | 0.23 |
ENSDART00000062378
|
adamts15b
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15b |
| chr5_-_50600512 | 0.23 |
ENSDART00000190318
|
mctp1a
|
multiple C2 domains, transmembrane 1a |
| chr22_-_38819603 | 0.22 |
ENSDART00000104437
|
si:ch211-262h13.5
|
si:ch211-262h13.5 |
| chr7_+_32722227 | 0.22 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr5_-_62988463 | 0.22 |
ENSDART00000047143
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr19_+_9277327 | 0.21 |
ENSDART00000104623
ENSDART00000151164 |
si:rp71-15k1.1
|
si:rp71-15k1.1 |
| chr14_+_36628131 | 0.21 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr4_-_21851473 | 0.21 |
ENSDART00000019748
|
lin7a
|
lin-7 homolog A (C. elegans) |
| chr5_+_33070608 | 0.21 |
ENSDART00000135063
|
npffr2b
|
neuropeptide FF receptor 2b |
| chr10_+_26597990 | 0.20 |
ENSDART00000079187
|
fhl1b
|
four and a half LIM domains 1b |
| chr4_-_5913338 | 0.20 |
ENSDART00000183590
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr25_-_19486399 | 0.19 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr17_+_31914877 | 0.19 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr8_+_35212233 | 0.19 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr20_+_47196506 | 0.18 |
ENSDART00000153040
|
znf975
|
zinc finger protein 975 |
| chr19_+_11984725 | 0.17 |
ENSDART00000185960
|
spag1a
|
sperm associated antigen 1a |
| chr15_+_25489406 | 0.17 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr22_+_11867639 | 0.17 |
ENSDART00000144677
|
mras
|
muscle RAS oncogene homolog |
| chr24_+_13642126 | 0.17 |
ENSDART00000126769
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr2_-_51507540 | 0.16 |
ENSDART00000166605
ENSDART00000161093 |
pigrl2.3
|
polymeric immunoglobulin receptor-like 2.3 |
| chr8_+_37749263 | 0.16 |
ENSDART00000108556
ENSDART00000147942 |
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd2_foxd5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0006824 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.8 | 4.0 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.5 | 1.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.5 | 2.0 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.5 | 1.5 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.4 | 3.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 1.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.3 | 4.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.2 | 0.7 | GO:0043605 | allantoin catabolic process(GO:0000256) cellular amide catabolic process(GO:0043605) |
| 0.2 | 2.0 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 0.6 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.2 | 1.8 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 1.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.8 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 2.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 0.4 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 1.6 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.1 | 0.9 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 2.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.8 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 3.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 1.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 3.1 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.8 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 2.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.6 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.1 | 0.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 3.0 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 3.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.1 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.5 | GO:0032095 | regulation of response to food(GO:0032095) |
| 0.1 | 0.5 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 3.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.8 | GO:0034198 | negative regulation of autophagy(GO:0010507) cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.1 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.6 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 1.7 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.6 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 0.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.3 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.9 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0042359 | response to vitamin D(GO:0033280) vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.6 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 2.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.6 | GO:0072376 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 2.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.4 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.1 | 0.4 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 2.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 2.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.9 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.2 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 2.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 2.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 3.0 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.5 | 1.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.4 | 4.0 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.4 | 2.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.6 | GO:0070643 | vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.3 | 6.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 2.8 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 1.7 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 2.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 1.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.2 | 1.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 2.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.2 | 2.7 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 2.0 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 0.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 3.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 2.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.7 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 5.1 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 0.4 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 3.0 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.4 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 3.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 1.1 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.5 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.8 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.9 | REACTOME OPSINS | Genes involved in Opsins |
| 0.3 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 4.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.2 | 3.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.1 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 1.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |